| Entry |
|
|---|
| Name |
Glycine, serine and threonine metabolism |
|---|
| Description |
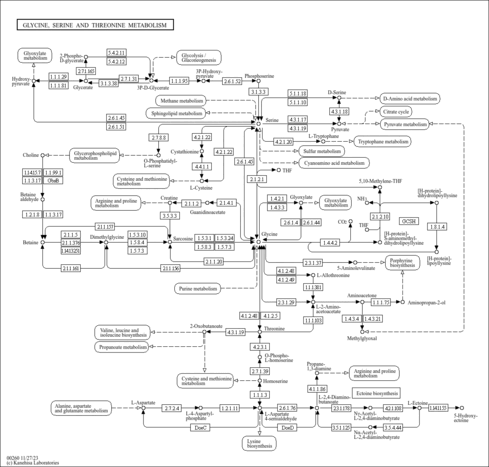
Serine is derived from 3-phospho-D-glycerate, an intermediate of glycolysis [MD: M00020], and glycine is derived from serine. Threonine is an essential amino acid, which animals cannot synthesize. In bacteria and plants, threonine is derived from aspartate [MD: M00018].
|
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
| rn00260 | Glycine, serine and threonine metabolism |
|
|---|
| Module |
| M00018 | Threonine biosynthesis, aspartate => homoserine => threonine [PATH:rn00260] |
| M00020 | Serine biosynthesis, glycerate-3P => serine [PATH:rn00260] |
| M00338 | Cysteine biosynthesis, homocysteine + serine => cysteine [PATH:rn00260] |
| M00974 | Betaine metabolism, animals, betaine => glycine [PATH:rn00260] |
| M00975 | Betaine degradation, bacteria, betaine => pyruvate [PATH:rn00260] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00364 | glycine:ferricytochrome-c oxidoreductase (deaminating) |
| R00366 | glycine:oxygen oxidoreductase (deaminating); aminoacetic acid:oxygen oxidoreductase (deaminating) |
| R00367 | S-adenosyl-L-methionine:glycine N-methyltransferase |
| R00371 | acetyl-CoA:glycine C-acetyltransferase |
| R00372 | glycine:2-oxoglutarate aminotransferase |
| R00480 | ATP:L-aspartate 4-phosphotransferase |
| R00565 | L-arginine:glycine amidinotransferase |
| R00582 | O-phospho-L-serine phosphohydrolase |
| R00585 | L-serine:pyruvate aminotransferase |
| R00588 | L-serine:glyoxylate aminotransferase |
| R00610 | sarcosine:oxygen oxidoreductase (demethylating) |
| R00611 | sarcosine:electron-transfer flavoprotein oxidoreductase (demethylating) |
| R00751 | L-threonine acetaldehyde-lyase (glycine-forming) |
| R00830 | succinyl-CoA:glycine C-succinyltransferase (decarboxylating) |
| R00891 | L-serine hydro-lyase (adding hydrogen sulfide, L-cysteine-forming) |
| R00945 | 5,10-methylenetetrahydrofolate:glycine hydroxymethyltransferase |
| R00996 | L-threonine ammonia-lyase (2-oxobutanoate-forming) |
| R01001 | L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming) |
| R01022 | choline:oxygen 1-oxidoreductase |
| R01025 | choline:acceptor 1-oxidoreductase |
| R01290 | L-serine hydro-lyase (adding homocysteine; L-cystathionine-forming) |
| R01388 | D-glycerate:NAD+ 2-oxidoreductase |
| R01392 | D-glycerate:NADP+ 2-oxidoreductase |
| R01465 | L-threonine:NAD+ oxidoreductase |
| R01466 | O-phospho-L-homoserine phosphate-lyase (adding water;L-threonine-forming) |
| R01511 | D-glycerate-3-phosphate phosphohydrolase |
| R01513 | 3-phospho-D-glycerate:NAD+ 2-oxidoreductase |
| R01514 | ATP:(R)-glycerate 3-phosphotransferase |
| R01518 | D-phosphoglycerate 2,3-phosphomutase |
| R01564 | N,N-dimethylglycine,5,6,7,8-tetrahydrofolate:oxygen oxidoreductase (demethylating,5,10-methylenetetrahydrofolate-forming) |
| R01565 | N,N-dimethylglycine,5,6,7,8-tetrahydrofolate:electron-transferflavoprotein oxidoreductase (demethylating,5,10-methylenetetrahydrofolate-forming) |
| R01566 | creatine amidinohydrolase |
| R01771 | ATP:L-homoserine O-phosphotransferase |
| R01773 | L-homoserine:NAD+ oxidoreductase |
| R01775 | L-homoserine:NADP+ oxidoreductase |
| R01800 | CDP-diacylglycerol:L-serine 3-phosphatidyltransferase |
| R01883 | S-adenosyl-L-methionine:guanidinoacetate N-methyltransferase |
| R02291 | L-aspartate-4-semialdehyde:NADP+ oxidoreductase (phosphorylating) |
| R02529 | aminoacetone:oxygen oxidoreductase(deaminating) |
| R02565 | p-cumic alcohol:NAD+ oxidoreductase |
| R02566 | p-cumic alcohol:NADP+ oxidoreductase |
| R02722 | L-serine hydro-lyase [adding 1-C-(indol-3-yl)glycerol 3-phosphate; L-tryptophan and glyceraldehyde-3-phosphate-forming] |
| R02821 | trimethylaminoacetate:L-homosysteine S-methyltransferase |
| R03425 | glycine:lipoylprotein oxidoreductase (decarboxylating and acceptor-aminomethylating) |
| R03759 | (R)-1-aminopropan-2-ol:NAD+ oxidoreductase |
| R03815 | dihydrolipoylprotein:NAD+ oxidoreductase |
| R04125 | [protein]-S8-aminomethyldihydrolipoyllysine:tetrahydrofolate aminomethyltransferase (ammonia-forming) |
| R04173 | 3-phosphoserine:2-oxoglutarate aminotransferase |
| R06171 | L-allo-threonine acetaldehyde-lyase (glycine-forming) |
| R06977 | L-2,4-diaminobutyrate:2-oxoglutarate 4-aminotransferase |
| R06978 | acetyl-CoA:L-2,4-diaminobutanoate N4-acetyltransferase |
| R07244 | S-adenosyl-L-methionine:N,N-dimethylglycine N-methyltransferase |
| R07409 | choline,reduced-ferredoxin:oxygen oxidoreductase |
| R07650 | L-2,4-diaminobutanoate carboxy-lyase |
| R08211 | betaine aldehyde:oxygen oxidoreductase |
| R08572 | ATP:(R)-glycerate 2-phosphotransferase |
| R09801 | N2-acetyl-L-2,4-diaminobutanoate amidohydrolase |
| R09805 | L-aspartate-4-semialdehyde:NAD+ oxidoreductase |
| R10060 | S-adenosyl-L-methionine:glycine N-methyltransferase [N,N-dimethylglycine-forming] |
| R10061 | S-adenosyl-L-methionine:sarcosine N-methyltransferase [betaine-forming] |
| R10851 | L-allo-threonine:NADP+ 3-oxidoreductase |
| R12787 | glycine betaine:[Co(I) glycine betaine-specific corrinoid protein] Co-methyltransferase |
| R12967 | sarcosine,5,6,7,8-tetrahydrofolate:O2 oxidoreductase (demethylating,5,10-methylenetetrahydrofolate-forming) |
| R13016 | glycine betaine,NADH:oxygen oxidoreductase (demethylating) |
| R13028 | N,N-dimethylglycine:ferredoxin oxidoreductase (demethylating) |
| R13029 | sarcosine:ferredoxin oxidoreductase (demethylating) |
|
|---|
| Compound |
| C00143 | 5,10-Methylenetetrahydrofolate |
| C00441 | L-Aspartate 4-semialdehyde |
| C01242 | [Protein]-S8-aminomethyldihydrolipoyllysine |
| C03508 | L-2-Amino-3-oxobutanoic acid |
| C06442 | N(gamma)-Acetyldiaminobutyrate |
| C19929 | N(alpha)-Acetyl-L-2,4-diaminobutyrate |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Kuhlmann AU, Bremer E. |
|---|
| Title |
Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Canovas D, Borges N, Vargas C, Ventosa A, Nieto JJ, Santos H. |
|---|
| Title |
Role of Ngamma-acetyldiaminobutyrate as an enzyme stabilizer and an intermediate in the biosynthesis of hydroxyectoine. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Bursy J, Pierik AJ, Pica N, Bremer E. |
|---|
| Title |
Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Garcia-Estepa R, Argandona M, Reina-Bueno M, Capote N, Iglesias-Guerra F, Nieto JJ, Vargas C. |
|---|
| Title |
The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Arevalo-Rodriguez M, Pan X, Boeke JD, Heitman J |
|---|
| Title |
FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rees WD, Hay SM |
|---|
| Title |
The biosynthesis of threonine by mammalian cells: expression of a complete bacterial biosynthetic pathway in an animal cell. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Jacques SL, Nieman C, Bareich D, Broadhead G, Kinach R, Honek JF, Wright GD |
|---|
| Title |
Characterization of yeast homoserine dehydrogenase, an antifungal target: the invariant histidine 309 is important for enzyme integrity. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chen X, Jhee KH, Kruger WD |
|---|
| Title |
Production of the neuromodulator H2S by cystathionine beta-synthase via the condensation of cysteine and homocysteine. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00250 | Alanine, aspartate and glutamate metabolism |
| rn00270 | Cysteine and methionine metabolism |
| rn00290 | Valine, leucine and isoleucine biosynthesis |
| rn00330 | Arginine and proline metabolism |
| rn00564 | Glycerophospholipid metabolism |
| rn00630 | Glyoxylate and dicarboxylate metabolism |
|
|---|
| KO pathway |
|
|---|