| Entry |
|
|---|
| Name |
Midecamycin (JP18/INN);
Medemycin (TN) |
|---|
| Formula |
C41H67NO15
|
|---|
| Exact mass |
813.4511
|
|---|
| Mol weight |
813.97
|
|---|
| Structure |
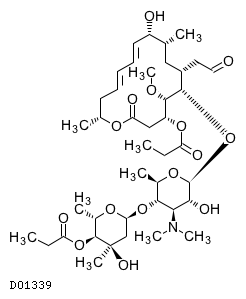
|
|---|
| Simcomp |
|
|---|
| Source |
Streptomyces mycarofaciens [TAX: 1949]
|
|---|
| Class |
Antibacterial
DG01551 Macrolide antibiotic
DG01876 16-Membered ring macrolide antibiotic
Metabolizing enzyme inhibitor
DG02852 CYP3A/CYP3A4 inhibitor
DG01522 CYP3A4 inhibitor
|
|---|
| Remark |
|
|---|
| Efficacy |
Antibacterial, Protein biosynthesis inhibitor |
|---|
| Target |
50S ribosomal subunit |
|---|
| Pathway |
|
|---|
| Interaction |
CYP inhibition: CYP3A4 [HSA: 1576]
|
|---|
| Structure map |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
J ANTIINFECTIVES FOR SYSTEMIC USE
J01 ANTIBACTERIALS FOR SYSTEMIC USE
J01F MACROLIDES, LINCOSAMIDES AND STREPTOGRAMINS
J01FA Macrolides
J01FA03 Midecamycin
D01339 Midecamycin (JP18/INN)
Drug groups [BR:br08330]
Antibacterial
DG01551 Macrolide antibiotic
DG01876 16-Membered ring macrolide antibiotic
DG01903 Midecamycin
D01339 Midecamycin
Metabolizing enzyme inhibitor
DG02852 CYP3A/CYP3A4 inhibitor
DG01522 CYP3A4 inhibitor
DG01903 Midecamycin
D01339 Midecamycin
Antimicrobials [BR:br08307]
Antibacterials
Protein biosynthesis inhibitor
Macrolide
D01339 Midecamycin (JP18/INN)
Drugs listed in the Japanese Pharmacopoeia [BR:br08311]
Chemicals
D01339 Midecamycin
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D01339
Drug groups [BR:br08330]
Antibacterial
DG01551 Macrolide antibiotic
DG01876 16-Membered ring macrolide antibiotic
DG01903 Midecamycin
Metabolizing enzyme inhibitor
DG02852 CYP3A/CYP3A4 inhibitor
DG01522 CYP3A4 inhibitor
DG01903 Midecamycin
Antimicrobials abbreviations [BR:br08327]
Antibacterials
Protein biosynthesis inhibitor
Macrolide
DG01903 Midecamycin
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 57
1 C1y C 26.8994 -21.1901
2 C1y C 27.9964 -21.8318
3 O2a O 25.8069 -21.8318
4 C1y C 26.8994 -19.9298
5 C1y C 29.1054 -21.1901
6 N1c N 27.9964 -23.0988
7 C1y C 24.7156 -22.4624
8 O2x O 27.9964 -19.2992
9 C1a C 25.8126 -19.2992
10 C1y C 29.1054 -19.9298
11 O1a O 30.2035 -21.8318
12 C1a C 29.0942 -23.7283
13 C1a C 26.9051 -23.7283
14 C1x C 24.7156 -23.7227
15 O2x O 23.6064 -21.8318
16 O2a O 31.9933 -16.8787
17 C1z C 23.6064 -24.3644
18 C1y C 22.5094 -22.4624
19 C1y C 28.5430 -13.4567
20 C1y C 22.5094 -23.7227
21 C1a C 22.7511 -25.5744
22 O1a O 24.4965 -25.7321
23 C1a C 21.4226 -21.8318
24 C1y C 28.5374 -12.1563
25 C1y C 27.4228 -14.1154
26 O7a O 21.4226 -24.3644
27 C1x C 27.4059 -11.5201
28 C1b C 29.6231 -11.5144
29 C1y C 27.4283 -15.4157
30 O2a O 26.4591 -13.4903
31 C1y C 27.4059 -10.2432
32 C4a C 30.7268 -12.1396
33 C1x C 26.2968 -16.0688
34 O7a O 28.5319 -16.0453
35 C1y C 26.2911 -9.6125
36 C1a C 28.4973 -9.6070
37 O4a O 31.8181 -11.4977
38 C7x C 25.1482 -15.4326
39 C2x C 25.1998 -10.2432
40 O1a O 26.2911 -8.3531
41 O7x O 24.0011 -16.0632
42 O6a O 25.1482 -14.1657
43 C2x C 25.1998 -11.5201
44 C1y C 22.8696 -15.3755
45 C2x C 24.0794 -12.1563
46 C1x C 22.9229 -14.0359
47 C1a C 21.7548 -15.9837
48 C2x C 24.0625 -13.4399
49 C1a C 26.4489 -12.2292
50 C7a C 20.3298 -23.7444
51 C1b C 19.2429 -24.3861
52 O6a O 20.3172 -22.4894
53 C1a C 18.1501 -23.7729
54 C7a C 28.5895 -17.3762
55 C1b C 27.3552 -18.0339
56 O6a O 29.6712 -18.0556
57 C1a C 26.2036 -17.3546
BOND 59
1 5 11 1 #Down
2 6 12 1
3 6 13 1
4 7 14 1
5 7 15 1
6 10 16 1 #Up
7 14 17 1
8 15 18 1
9 19 16 1 #Down
10 17 20 1
11 17 21 1 #Down
12 17 22 1 #Up
13 18 23 1 #Down
14 19 24 1
15 19 25 1
16 20 26 1 #Up
17 24 27 1
18 24 28 1 #Down
19 25 29 1
20 25 30 1 #Down
21 27 31 1
22 28 32 1
23 29 33 1
24 29 34 1 #Down
25 31 35 1
26 31 36 1 #Down
27 32 37 2
28 33 38 1
29 35 39 1
30 35 40 1 #Down
31 38 41 1
32 38 42 2
33 39 43 2
34 41 44 1
35 43 45 1
36 44 46 1
37 44 47 1 #Down
38 45 48 2
39 8 10 1
40 18 20 1
41 46 48 1
42 30 49 1
43 1 2 1
44 26 50 1
45 1 3 1 #Down
46 50 51 1
47 1 4 1
48 50 52 2
49 2 5 1
50 51 53 1
51 2 6 1 #Up
52 7 3 1 #Up
53 34 54 1
54 4 8 1
55 54 55 1
56 4 9 1 #Up
57 54 56 2
58 5 10 1
59 55 57 1
|
|---|