| Entry |
|
|---|
| Name |
Clemastine (USAN) |
|---|
| Formula |
C21H26ClNO
|
|---|
| Exact mass |
343.1703
|
|---|
| Mol weight |
343.89
|
|---|
| Structure |
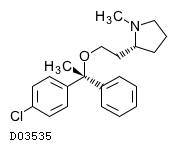
|
|---|
| Simcomp |
|
|---|
| Class |
Anti-allergic agent
DG01557 Histamine receptor antagonist
DG01482 Histamine receptor H1 antagonist
Metabolizing enzyme inhibitor
DG01645 CYP2D6 inhibitor
|
|---|
| Remark |
| Product (DG00387): | D00666<JP/US> |
|
|---|
| Efficacy |
Antiallergic, H1 receptor antagonist |
|---|
| Comment |
Diphenhydramine derivative
|
|---|
| Target |
|
|---|
| Pathway |
| hsa04080 | Neuroactive ligand-receptor interaction |
|
|---|
| Interaction |
CYP inhibition: CYP2D6 [HSA: 1565]
|
|---|
| Structure map |
| map07212 | Histamine H1 receptor antagonists |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
D DERMATOLOGICALS
D04 ANTIPRURITICS, INCL. ANTIHISTAMINES, ANESTHETICS, ETC.
D04A ANTIPRURITICS, INCL. ANTIHISTAMINES, ANESTHETICS, ETC.
D04AA Antihistamines for topical use
D04AA14 Clemastine
D03535 Clemastine (USAN)
R RESPIRATORY SYSTEM
R06 ANTIHISTAMINES FOR SYSTEMIC USE
R06A ANTIHISTAMINES FOR SYSTEMIC USE
R06AA Aminoalkyl ethers
R06AA04 Clemastine
D03535 Clemastine (USAN)
Risk category of Japanese OTC drugs [BR:br08312]
Second-class OTC drugs
Inorganic and organic chemicals
Clemastine
D03535 Clemastine (USAN)
Drug groups [BR:br08330]
Anti-allergic agent
DG01557 Histamine receptor antagonist
DG01482 Histamine receptor H1 antagonist
DG00387 Clemastine
D03535 Clemastine
Metabolizing enzyme inhibitor
DG01645 CYP2D6 inhibitor
DG00387 Clemastine
D03535 Clemastine
Target-based classification of drugs [BR:br08310]
G Protein-coupled receptors
Rhodopsin family
Histamine
HRH1
D03535 Clemastine (USAN)
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D03535
Drug groups [BR:br08330]
Anti-allergic agent
DG01557 Histamine receptor antagonist
DG01482 Histamine receptor H1 antagonist
DG00387 Clemastine
Metabolizing enzyme inhibitor
DG01645 CYP2D6 inhibitor
DG00387 Clemastine
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 24
1 C1y C 12.6951 -14.0046
2 N1y N 12.6951 -12.6046
3 C1a C 11.4619 -11.8926
4 C1x C 14.0266 -14.4373
5 C1x C 14.8495 -13.3046
6 C1x C 14.0266 -12.1720
7 C8x C 5.4600 -16.8000
8 C8y C 5.4600 -18.2000
9 C8x C 6.6724 -18.9000
10 C8x C 7.8849 -18.2000
11 C8y C 7.8849 -16.8000
12 C8x C 6.6724 -16.1000
13 C1d C 9.0973 -16.1000
14 X Cl 4.2476 -18.9000
15 O2a O 9.0973 -14.7000
16 C1b C 10.3118 -13.9988
17 C1b C 11.5083 -14.6898
18 C8x C 10.3097 -18.2000
19 C8y C 10.3097 -16.8000
20 C8x C 11.5222 -18.9000
21 C8x C 12.7346 -18.2000
22 C8x C 12.7346 -16.8000
23 C8x C 11.5222 -16.1000
24 C1a C 7.8849 -15.4000
BOND 26
1 1 2 1
2 2 3 1
3 1 4 1
4 4 5 1
5 5 6 1
6 2 6 1
7 7 8 2
8 8 9 1
9 9 10 2
10 10 11 1
11 11 12 2
12 7 12 1
13 11 13 1
14 8 14 1
15 13 15 1
16 15 16 1
17 16 17 1
18 18 19 1
19 18 20 2
20 20 21 1
21 21 22 2
22 22 23 1
23 19 23 2
24 13 19 1 #Down
25 13 24 1 #Up
26 1 17 1 #Down
|
|---|