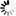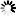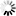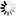2-Oxocarboxylic acids, also called 2-oxo acids and alpha-keto acids, are the most elementary set of metabolites that includes pyruvate (2-oxopropanoate), 2-oxobutanoate, oxaloacetate (2-oxosuccinate) and 2-oxoglutarate. This diagram illustrates the architecture of chain extension and modification reaction modules for 2-oxocarboxylic acids. The chain extension module
RM001 is a tricarboxylic pathway where acetyl-CoA derived carbon is used to extend the chain length by one. The chain modification modules
RM002 (including
RM032) and
RM033, together with a reductive amination step (
RC00006 or
RC00036), generate basic and branched-chain amino acids, respectively. The modification module
RM030 is used in the biosynthesis of glucosinolates, a class of plant secondary metabolites, for conversion to oxime followed by addition of thio-glucose moiety. Furthermore, the chain extension from 2-oxoadipate to 2-oxosuberate is followed by coenzyme B biosynthesis in methonogenic archaea.
 2-Oxocarboxylic acid metabolism - Ornithorhynchus anatinus (platypus)
2-Oxocarboxylic acid metabolism - Ornithorhynchus anatinus (platypus)
 2-Oxocarboxylic acid metabolism - Ornithorhynchus anatinus (platypus)
2-Oxocarboxylic acid metabolism - Ornithorhynchus anatinus (platypus)