 | | PATHWAY: spu01240 | |
| Entry |
|
|---|
| Name |
Biosynthesis of cofactors - Strongylocentrotus purpuratus (purple sea urchin) |
|---|
| Class |
|
|---|
| Pathway map |
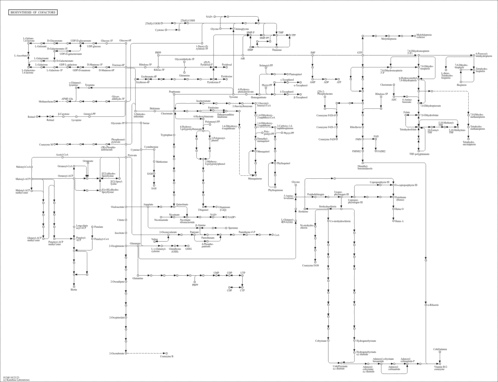
|
|---|
| Module |
| spu_M00128 | Ubiquinone biosynthesis, eukaryotes, 4-hydroxybenzoate + polyprenyl-PP => ubiquinol [PATH:spu01240] |
| spu_M00882 | Lipoic acid biosynthesis, eukaryotes, octanoyl-ACP => dihydrolipoyl-H [PATH:spu01240] |
| spu_M00883 | Lipoic acid biosynthesis, animals and bacteria, octanoyl-ACP => dihydrolipoyl-H => dihydrolipoyl-E2 [PATH:spu01240] |
|
|---|
| Organism |
Strongylocentrotus purpuratus (purple sea urchin) [GN: spu]
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|