| Entry |
|
|---|
| Name |
Phenylalanine metabolism - Streptomyces sp. SirexAA-E |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
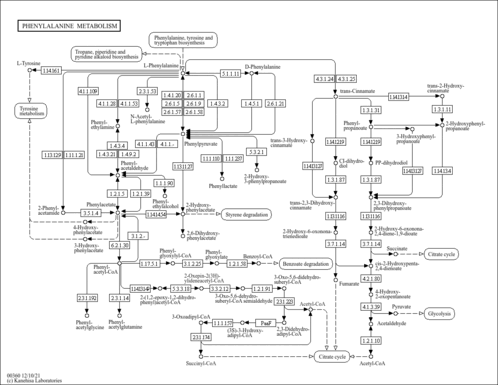
|
|---|
| Other DBs |
|
|---|
| Organism |
Streptomyces sp. SirexAA-E [GN: ssx]
|
|---|
| Gene |
|
|---|
| Compound |
| C00596 | 2-Hydroxy-2,4-pentadienoate |
| C01198 | 3-(2-Hydroxyphenyl)propanoate |
| C01772 | trans-2-Hydroxycinnamate |
| C02137 | alpha-Oxo-benzeneacetic acid |
| C02763 | 2-Hydroxy-3-phenylpropenoate |
| C03519 | N-Acetyl-L-phenylalanine |
| C03589 | 4-Hydroxy-2-oxopentanoate |
| C04044 | 3-(2,3-Dihydroxyphenyl)propanoate |
| C04479 | 2-Hydroxy-6-oxonona-2,4-diene-1,9-dioate |
| C06207 | 2,6-Dihydroxyphenylacetate |
| C11457 | 3-(3-Hydroxyphenyl)propanoic acid |
| C11588 | cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol |
| C12621 | trans-3-Hydroxycinnamate |
| C12622 | cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol |
| C12623 | trans-2,3-Dihydroxycinnamate |
| C12624 | 2-Hydroxy-6-ketononatrienedioate |
| C14144 | 5-Carboxy-2-pentenoyl-CoA |
| C14145 | (3S)-3-Hydroxyadipyl-CoA |
| C19945 | 3-Oxo-5,6-dehydrosuberyl-CoA |
| C19946 | 3-Oxo-5,6-dehydrosuberyl-CoA semialdehyde |
| C19975 | 2-Oxepin-2(3H)-ylideneacetyl-CoA |
| C20062 | 2-(1,2-Epoxy-1,2-dihydrophenyl)acetyl-CoA |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Arai H, Yamamoto T, Ohishi T, Shimizu T, Nakata T, Kudo T. |
|---|
| Title |
Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Diaz E, Ferrandez A, Garcia JL. |
|---|
| Title |
Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ferrandez A, Garcia JL, Diaz E. |
|---|
| Title |
Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hwang JY, Park J, Seo JH, Cha M, Cho BK, Kim J, Kim BG |
|---|
| Title |
Simultaneous synthesis of 2-phenylethanol and L-homophenylalanine using aromatic transaminase with yeast Ehrlich pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G |
|---|
| Title |
Bacterial phenylalanine and phenylacetate catabolic pathway revealed. |
|---|
| Journal |
|
|---|
Related
pathway |
| ssx00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|