| Entry |
|
|---|
| Name |
Levomethadyl acetate (USAN);
Levacetylmethadol (INN) |
|---|
| Formula |
C23H31NO2
|
|---|
| Exact mass |
353.2355
|
|---|
| Mol weight |
353.50
|
|---|
| Structure |
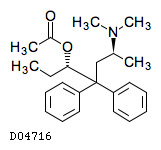
|
|---|
| Simcomp |
|
|---|
| Class |
Neuropsychiatric agent
DG01564 Opioid receptor agonist
DG01563 mu-Opioid receptor agonist
Analgesic
DG01984 Opioid analgesics
Metabolizing enzyme substrate
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
|
|---|
| Remark |
|
|---|
| Efficacy |
Analgesic (narcotic), Opioid receptor agonist |
|---|
| Target |
|
|---|
| Pathway |
| hsa04080 | Neuroactive ligand-receptor interaction |
|
|---|
| Metabolism |
Enzyme: CYP3A4 [HSA: 1576]
|
|---|
| Interaction |
|
|---|
| Structure map |
| map07224 | Opioid receptor agonists/antagonists |
|
|---|
| Other map |
| map00982 | Drug metabolism - cytochrome P450 |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
N NERVOUS SYSTEM
N07 OTHER NERVOUS SYSTEM DRUGS
N07B DRUGS USED IN ADDICTIVE DISORDERS
N07BC Drugs used in opioid dependence
N07BC03 Levacetylmethadol
D04716 Levomethadyl acetate (USAN)
Drug groups [BR:br08330]
Neuropsychiatric agent
DG01564 Opioid receptor agonist
DG01563 mu-Opioid receptor agonist
DG01000 Levacetylmethadol
D04716 Levomethadyl acetate
Analgesic
DG01984 Opioid analgesics
DG01000 Levacetylmethadol
D04716 Levomethadyl acetate
Metabolizing enzyme substrate
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
DG01000 Levacetylmethadol
D04716 Levomethadyl acetate
Target-based classification of drugs [BR:br08310]
G Protein-coupled receptors
Rhodopsin family
Opioid
OPRM1
D04716 Levomethadyl acetate (USAN)
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D04716
Drug groups [BR:br08330]
Neuropsychiatric agent
DG01564 Opioid receptor agonist
DG01563 mu-Opioid receptor agonist
DG01000 Levacetylmethadol
Analgesic
DG01984 Opioid analgesics
DG01000 Levacetylmethadol
Metabolizing enzyme substrate
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
DG01000 Levacetylmethadol
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 26
1 C8x C 21.8383 -21.8142
2 C8x C 21.8383 -23.1473
3 C8x C 23.0311 -23.8490
4 C8x C 24.2239 -23.1473
5 C8y C 24.2239 -21.8142
6 C8x C 23.0311 -21.1125
7 C8x C 26.6095 -23.1473
8 C8y C 26.6095 -21.8142
9 C1d C 25.4167 -21.1125
10 C8x C 27.7321 -23.8490
11 C8x C 28.9249 -23.1473
12 C8x C 28.9249 -21.8142
13 C8x C 27.7321 -21.1125
14 C1b C 25.4167 -19.7092
15 C1c C 26.6095 -19.0778
16 C1c C 23.3819 -19.9899
17 C1a C 27.7321 -19.7092
18 N1c N 26.6095 -17.6745
19 C1a C 27.7321 -16.9728
20 C1a C 25.3465 -16.9728
21 O7a O 23.3819 -18.6568
22 C1b C 22.1891 -20.6916
23 C1a C 21.0665 -19.9899
24 C7a C 22.1891 -17.9551
25 C1a C 20.9963 -18.6568
26 O6a O 22.1891 -16.5518
BOND 27
1 1 2 2
2 2 3 1
3 3 4 2
4 4 5 1
5 5 6 2
6 1 6 1
7 7 8 1
8 8 9 1
9 5 9 1
10 7 10 2
11 10 11 1
12 11 12 2
13 12 13 1
14 8 13 2
15 9 14 1
16 14 15 1
17 9 16 1
18 15 17 1
19 15 18 1 #Up
20 18 19 1
21 18 20 1
22 16 21 1 #Down
23 16 22 1
24 22 23 1
25 21 24 1
26 24 25 1
27 24 26 2
|
|---|