| Entry |
|
|---|
| Name |
Dordaviprone hydrochloride (JAN/USAN);
Dordaviprone dihydrochloride;
Modeyso (TN) |
|---|
| Product |
|
|---|
| Formula |
C24H26N4O. 2HCl
|
|---|
| Exact mass |
458.1640
|
|---|
| Mol weight |
459.41
|
|---|
| Structure |
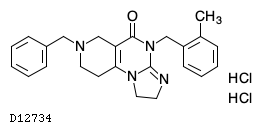
|
|---|
| Class |
|
|---|
| Remark |
| Product (DG03280): | D12734<US> |
|
|---|
| Efficacy |
Antineoplastic |
|---|
| Disease |
Diffuse midline glioma (H3 K27M mutation) |
|---|
| Target |
|
|---|
| Metabolism |
Enzyme: CYP3A4 [HSA: 1576]; CYP2B6 [HSA: 1555], CYP2C8 [HSA: 1558], CYP2C9 [HSA: 1559], CYP2D6 [HSA: 1565], CYP3A5 [HSA: 1577]
|
|---|
| Interaction |
|
|---|
| Brite |
Drug groups [BR:br08330]
Metabolizing enzyme substrate
DG02919 CYP2B6 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
DG02918 CYP2C8 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
DG01642 CYP2C9 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
DG01644 CYP2D6 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
DG02925 CYP3A5 substrate
DG03280 Dordaviprone
D12734 Dordaviprone hydrochloride
Target-based classification of drugs [BR:br08310]
G Protein-coupled receptors
Rhodopsin family
Dopamine
DRD2
D12734 Dordaviprone hydrochloride (JAN/USAN) <US>
Peptidases and inhibitors
Serine peptidases
ClpP endopeptidase family
CLPP
D12734 Dordaviprone hydrochloride (JAN/USAN) <US>
New drug approvals in the USA [br08319.html]
New molecular entities and new therapeutic biological products
D12734
New drug approvals in the USA, Europe and Japan [br08328.html]
Approval dates by FDA, EMA and PMDA
D12734
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D12734
Drug groups [BR:br08330]
Metabolizing enzyme substrate
DG02919 CYP2B6 substrate
DG03280 Dordaviprone
DG02918 CYP2C8 substrate
DG03280 Dordaviprone
DG01642 CYP2C9 substrate
DG03280 Dordaviprone
DG01644 CYP2D6 substrate
DG03280 Dordaviprone
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
DG03280 Dordaviprone
DG02925 CYP3A5 substrate
DG03280 Dordaviprone
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 31
1 C8y C 18.6602 -16.5636
2 C8y C 18.6602 -15.1658
3 C8y C 19.8483 -14.4669
4 N4y N 21.1063 -15.1658
5 C8y C 21.1063 -16.5636
6 N4y N 19.8483 -17.2625
7 C1x C 17.4721 -17.2625
8 C1x C 16.2141 -16.5636
9 N1y N 16.2141 -15.1658
10 C1x C 17.4721 -14.4669
11 C1b C 15.0260 -14.4669
12 C8y C 13.8379 -15.1658
13 C8x C 13.8379 -16.5636
14 C8x C 12.5799 -17.2625
15 C8x C 11.3918 -16.5636
16 C8x C 11.3918 -15.1658
17 C8x C 12.5799 -14.4669
18 O5x O 19.8483 -13.0692
19 C1b C 22.2944 -14.4669
20 C8y C 23.4825 -15.1658
21 C8y C 24.6706 -14.4669
22 C8x C 25.8587 -15.1658
23 C8x C 25.8587 -16.5636
24 C8x C 24.7405 -17.2625
25 C8x C 23.4825 -16.5636
26 C1a C 24.6706 -13.0692
27 N2x N 22.1547 -17.4721
28 C1x C 21.5257 -18.8000
29 C1x C 20.1279 -18.6602
30 X Cl 27.7200 -17.4300
31 X Cl 27.7200 -18.8300
BOND 33
1 1 2 2
2 2 3 1
3 3 4 1
4 4 5 1
5 5 6 1
6 1 6 1
7 1 7 1
8 7 8 1
9 8 9 1
10 9 10 1
11 2 10 1
12 9 11 1
13 11 12 1
14 12 13 2
15 13 14 1
16 14 15 2
17 15 16 1
18 16 17 2
19 12 17 1
20 3 18 2
21 4 19 1
22 19 20 1
23 20 21 2
24 21 22 1
25 22 23 2
26 23 24 1
27 24 25 2
28 20 25 1
29 21 26 1
30 5 27 2
31 27 28 1
32 28 29 1
33 6 29 1
|
|---|