| Entry |
|
|---|
| Name |
(R)-5-phosphomevalonate hydro-lyase
|
|---|
| Definition |
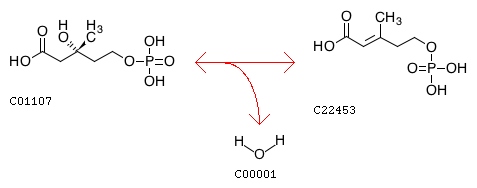
(R)-5-Phosphomevalonate <=> trans-Anhydromevalonate 5-phosphate + H2O
|
|---|
| Equation |
|
|---|
|
 |
|---|
| Comment |
mevalonate 5-phosphate dehydratase
|
|---|
| Reaction class |
|
|---|
| Enzyme |
|
|---|
| Pathway |
| rn00900 | Terpenoid backbone biosynthesis |
| rn01110 | Biosynthesis of secondary metabolites |
|
|---|
| Module |
| M00849 | C5 isoprenoid biosynthesis, mevalonate pathway, archaea |
|
|---|
| Brite |
Enzymatic reactions [BR:br08201]
4. Lyase reactions
4.2 Carbon-oxygen lyases
4.2.1 Hydro-lyases
4.2.1.182
R12894 (R)-5-Phosphomevalonate <=> trans-Anhydromevalonate 5-phosphate + H2O
|
|---|
| Orthology |
|
|---|
| Reference |
|
|---|
| Authors |
Hayakawa H, Motoyama K, Sobue F, Ito T, Kawaide H, Yoshimura T, Hemmi H. |
|---|
| Title |
Modified mevalonate pathway of the archaeon Aeropyrum pernix proceeds via trans-anhydromevalonate 5-phosphate. |
|---|
| Journal |
|
|---|
| LinkDB |
|
|---|