| Entry |
|
|---|
| Name |
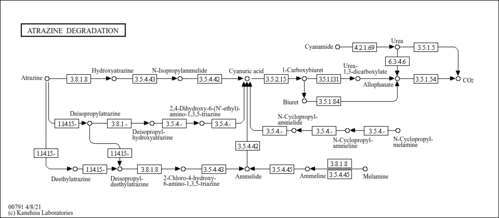
Atrazine degradation - Methylorubrum extorquens DM4 |
|---|
| Class |
Metabolism; Xenobiotics biodegradation and metabolism
|
|---|
| Pathway map |
|
|---|
| Other DBs |
|
|---|
| Organism |
Methylorubrum extorquens DM4 [GN: mdi]
|
|---|
| Gene |
| METDI1775 | ureAB; urease gamma (N-terminal) and beta (C-terminal) subunits (urea amidohydrolase gamma/beta subunits) [KO:K14048] [EC:3.5.1.5] |
| METDI3791 | ureAB; urease gamma (N-terminal) and beta (C-terminal) subunits (urea amidohydrolase gamma/beta subunits) [KO:K14048] [EC:3.5.1.5] |
| METDI4404 | ureAB; urease gamma (N-terminal) and beta (C-terminal) subunits (urea amidohydrolase gamma/beta subunits) [KO:K14048] [EC:3.5.1.5] |
|
|---|
| Compound |
| C06557 | Deisopropylhydroxyatrazine |
| C06560 | Deisopropyldeethylatrazine |
| C08735 | 2-Chloro-4-hydroxy-6-amino-1,3,5-triazine |
|
|---|
| Reference |
|
|---|
| Authors |
Shapir N, Cheng G, Sadowsky MJ, Wackett LP |
|---|
| Title |
Purification and characterization of TrzF: biuret hydrolysis by allophanate hydrolase supports growth. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cheng G, Shapir N, Sadowsky MJ, Wackett LP. |
|---|
| Title |
Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Li J, Biss M, Fu Y, Xu X, Moore SA, Xiao W |
|---|
| Title |
Two duplicated genes DDI2 and DDI3 in budding yeast encode a cyanamide hydratase and are induced by cyanamide. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|