| Entry |
|
|---|
| Name |
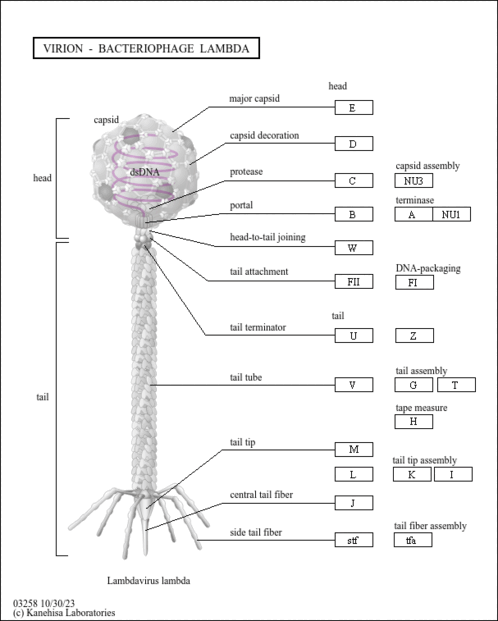
Virion - Bacteriophage lambda - Escherichia coli O45:K1:H7 S88 (ExPEC) |
|---|
| Class |
Genetic Information Processing; Information processing in viruses
|
|---|
| Pathway map |
|
|---|
| Organism |
Escherichia coli O45:K1:H7 S88 (ExPEC) [GN: ecz]
|
|---|
| Gene |
|
|---|
| Reference |
|
|---|
| Authors |
Rajagopala SV, Casjens S, Uetz P |
|---|
| Title |
The protein interaction map of bacteriophage lambda. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wang C, Zeng J, Wang J |
|---|
| Title |
Structural basis of bacteriophage lambda capsid maturation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cardarelli L, Pell LG, Neudecker P, Pirani N, Liu A, Baker LA, Rubinstein JL, Maxwell KL, Davidson AR |
|---|
| Title |
Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Campbell PL, Duda RL, Nassur J, Conway JF, Huet A |
|---|
| Title |
Mobile Loops and Electrostatic Interactions Maintain the Flexible Tail Tube of Bacteriophage Lambda. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu J, Hendrix RW, Duda RL |
|---|
| Title |
A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu J, Hendrix RW, Duda RL |
|---|
| Title |
Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hauser R, Blasche S, Dokland T, Haggard-Ljungquist E, von Brunn A, Salas M, Casjens S, Molineux I, Uetz P |
|---|
| Title |
Bacteriophage protein-protein interactions. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Goulet A, Spinelli S, Mahony J, Cambillau C |
|---|
| Title |
Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|