 | | PATHWAY: haso01232 | |
| Entry |
|
|---|
| Name |
Nucleotide metabolism - Halomonas sp. 'Soap Lake #7' |
|---|
| Class |
|
|---|
| Pathway map |
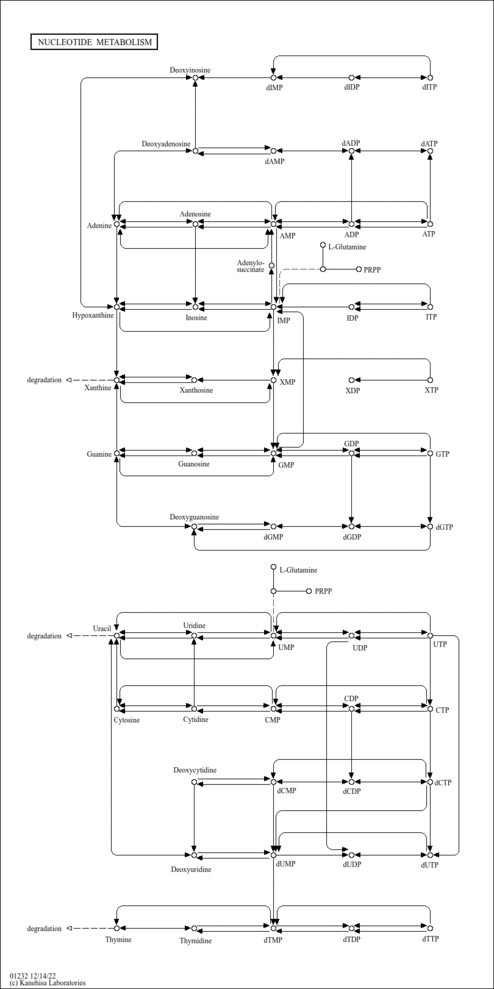
|
|---|
| Module |
| haso_M00053 | Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP [PATH:haso01232] |
|
|---|
| Organism |
Halomonas sp. 'Soap Lake #7' [GN: haso]
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|