 | | PATHWAY: hazt01100 | |
| Entry |
|
|---|
| Name |
Metabolic pathways - Hyalella azteca |
|---|
| Class |
|
|---|
| Pathway map |
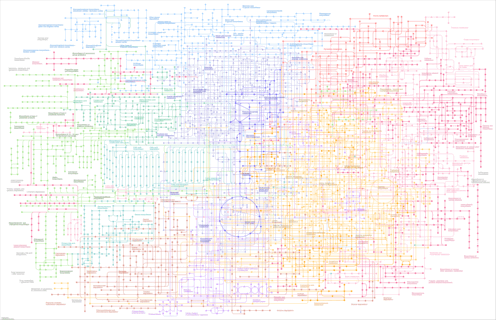
|
|---|
| Module |
| hazt_M00007 | Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P [PATH:hazt01100] |
| hazt_M00042 | Catecholamine biosynthesis, tyrosine => dopamine => noradrenaline => adrenaline [PATH:hazt01100] |
| hazt_M00046 | Pyrimidine degradation, uracil => beta-alanine, thymine => 3-aminoisobutanoate [PATH:hazt01100] |
| hazt_M00053 | Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP [PATH:hazt01100] |
| hazt_M00067 | Sulfoglycolipids biosynthesis, ceramide/1-alkyl-2-acylglycerol => sulfatide/seminolipid [PATH:hazt01100] |
| hazt_M00130 | Inositol phosphate metabolism, PI=> PIP2 => Ins(1,4,5)P3 => Ins(1,3,4,5)P4 [PATH:hazt01100] |
| hazt_M00143 | NADH dehydrogenase (ubiquinone) Fe-S protein/flavoprotein complex, mitochondria [PATH:hazt01100] |
| hazt_M00883 | Lipoic acid biosynthesis, animals and bacteria, octanoyl-ACP => dihydrolipoyl-H => dihydrolipoyl-E2 [PATH:hazt01100] |
| hazt_M00892 | UDP-N-acetyl-D-glucosamine biosynthesis, eukaryotes, glucose => UDP-GlcNAc [PATH:hazt01100] |
|
|---|
| Organism |
Hyalella azteca [GN: hazt]
|
|---|
Related
pathway |
| hazt00040 | Pentose and glucuronate interconversions |
| hazt00130 | Ubiquinone and other terpenoid-quinone biosynthesis |
| hazt00250 | Alanine, aspartate and glutamate metabolism |
| hazt00260 | Glycine, serine and threonine metabolism |
| hazt00280 | Valine, leucine and isoleucine degradation |
| hazt00290 | Valine, leucine and isoleucine biosynthesis |
| hazt00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| hazt00440 | Phosphonate and phosphinate metabolism |
| hazt00513 | Various types of N-glycan biosynthesis |
| hazt00514 | Other types of O-glycan biosynthesis |
| hazt00520 | Amino sugar and nucleotide sugar metabolism |
| hazt00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate |
| hazt00533 | Glycosaminoglycan biosynthesis - keratan sulfate |
| hazt00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin |
| hazt00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis |
| hazt00601 | Glycosphingolipid biosynthesis - lacto and neolacto series |
| hazt00603 | Glycosphingolipid biosynthesis - globo and isoglobo series |
| hazt00604 | Glycosphingolipid biosynthesis - ganglio series |
| hazt00630 | Glyoxylate and dicarboxylate metabolism |
| hazt00760 | Nicotinate and nicotinamide metabolism |
| hazt00980 | Metabolism of xenobiotics by cytochrome P450 |
| hazt01040 | Biosynthesis of unsaturated fatty acids |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|