 | | PATHWAY: mnt01240 | |
| Entry |
|
|---|
| Name |
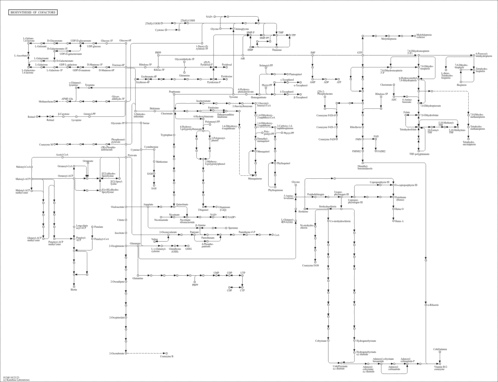
Biosynthesis of cofactors - Morus notabilis |
|---|
| Class |
|
|---|
| Pathway map |
|
|---|
| Module |
| mnt_M00112 | Tocopherol/tocotorienol biosynthesis, homogentisate + phytyl/geranylgeranyl-PP => tocopherol/tocotorienol [PATH:mnt01240] |
| mnt_M00125 | Riboflavin biosynthesis, plants and bacteria, GTP => riboflavin/FMN/FAD [PATH:mnt01240] |
| mnt_M00881 | Lipoic acid biosynthesis, plants and bacteria, octanoyl-ACP => dihydrolipoyl-E2/H [PATH:mnt01240] |
| mnt_M00882 | Lipoic acid biosynthesis, eukaryotes, octanoyl-ACP => dihydrolipoyl-H [PATH:mnt01240] |
| mnt_M00897 | Thiamine biosynthesis, plants, AIR (+ NAD+) => TMP/thiamine/TPP [PATH:mnt01240] |
| mnt_M00916 | Pyridoxal-P biosynthesis, R5P + glyceraldehyde-3P + glutamine => pyridoxal-P [PATH:mnt01240] |
| mnt_M00932 | Phylloquinone biosynthesis, chorismate (+ phytyl-PP) => phylloquinol [PATH:mnt01240] |
| mnt_M00933 | Plastoquinone biosynthesis, homogentisate + solanesyl-PP => plastoquinol [PATH:mnt01240] |
|
|---|
| Organism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|