| Entry |
|
|---|
| Name |
Basal transcription factors - Plutella xylostella (diamondback moth) |
|---|
| Class |
Genetic Information Processing; Transcription
|
|---|
| Pathway map |
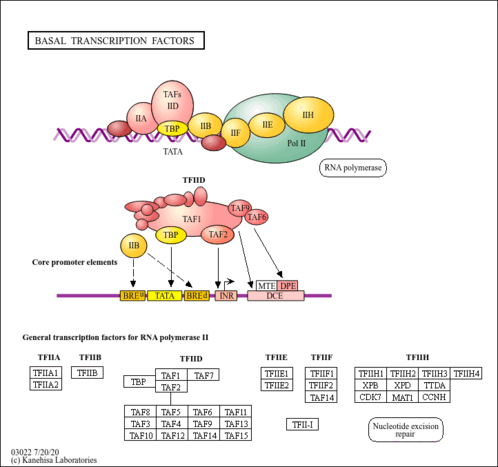
|
|---|
| Other DBs |
|
|---|
| Organism |
Plutella xylostella (diamondback moth) [GN: pxy]
|
|---|
| Gene |
| 105389183 | TAF5-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 5L isoform X1 [KO:K03130] |
| 105389688 | transcription initiation factor TFIID subunit 4 isoform X1 [KO:K03129] |
| 105387715 | transcription initiation factor TFIID subunit 6 isoform X1 [KO:K03131] |
| 105384461 | transcription initiation factor TFIID subunit 13 isoform X1 [KO:K03127] |
| 105389844 | general transcription factor IIE subunit 1 isoform X1 [KO:K03136] |
| 105385870 | general transcription factor IIF subunit 2 [KO:K03139] [EC:5.6.2.-] |
| 105389558 | general transcription factor IIH subunit 3 isoform X1 [KO:K03143] |
|
|---|
| Reference |
|
|---|
| Authors |
Thomas MC, Chiang CM |
|---|
| Title |
The general transcription machinery and general cofactors. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Martinez E |
|---|
| Title |
Multi-protein complexes in eukaryotic gene transcription. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cler E, Papai G, Schultz P, Davidson I |
|---|
| Title |
Recent advances in understanding the structure and function of general transcription factor TFIID. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Malik S, Roeder RG. |
|---|
| Title |
Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|