| Entry |
|
|---|
| Name |
Microbial metabolism in diverse environments - Rhodobiaceae bacterium SMS8 |
|---|
| Class |
|
|---|
| Pathway map |
| rbs01120 | Microbial metabolism in diverse environments |
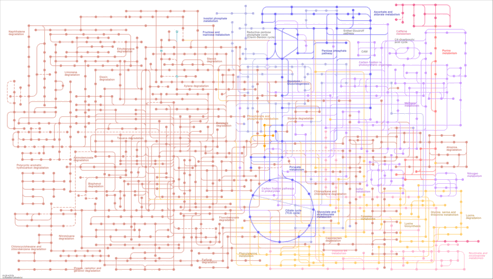
|
|---|
| Module |
| rbs_M00007 | Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P [PATH:rbs01120] |
|
|---|
| Organism |
Rhodobiaceae bacterium SMS8 [GN: rbs]
|
|---|
Related
pathway |
| rbs00051 | Fructose and mannose metabolism |
| rbs00053 | Ascorbate and aldarate metabolism |
| rbs00260 | Glycine, serine and threonine metabolism |
| rbs00361 | Chlorocyclohexane and chlorobenzene degradation |
| rbs00440 | Phosphonate and phosphinate metabolism |
| rbs00625 | Chloroalkane and chloroalkene degradation |
| rbs00630 | Glyoxylate and dicarboxylate metabolism |
| rbs00710 | Carbon fixation by Calvin cycle |
| rbs00760 | Nicotinate and nicotinamide metabolism |
| rbs00907 | Pinene, camphor and geraniol degradation |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|