 | | PATHWAY: salu01240 | |
| Entry |
|
|---|
| Name |
Biosynthesis of cofactors - Streptomyces noursei NK660 |
|---|
| Class |
|
|---|
| Pathway map |
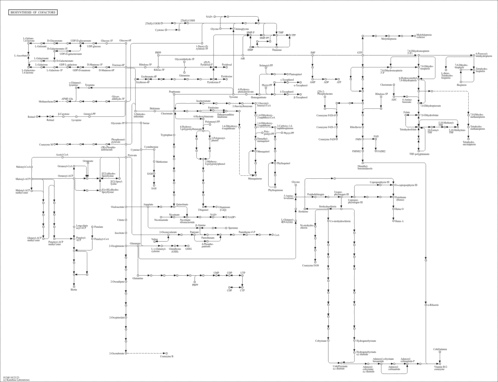
|
|---|
| Module |
| salu_M00881 | Lipoic acid biosynthesis, plants and bacteria, octanoyl-ACP => dihydrolipoyl-E2/H [PATH:salu01240] |
| salu_M00916 | Pyridoxal-P biosynthesis, R5P + glyceraldehyde-3P + glutamine => pyridoxal-P [PATH:salu01240] |
| salu_M00926 | Heme biosynthesis, bacteria, glutamyl-tRNA => coproporphyrin III => heme [PATH:salu01240] |
|
|---|
| Organism |
Streptomyces noursei NK660 [GN: salu]
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|