| Entry |
|
|---|
| Name |
beta-Alanine metabolism - Sinorhizobium sp. RAC02 |
|---|
| Class |
Metabolism; Metabolism of other amino acids
|
|---|
| Pathway map |
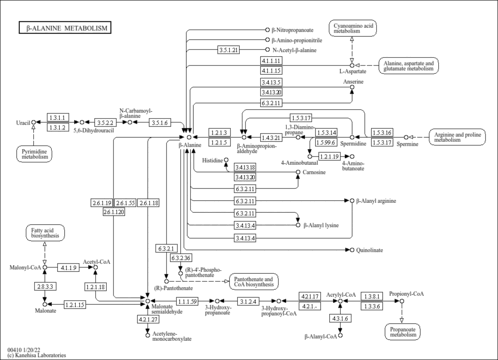
|
|---|
| Module |
| six_M00046 | Pyrimidine degradation, uracil => beta-alanine, thymine => 3-aminoisobutanoate [PATH:six00410] |
|
|---|
| Other DBs |
|
|---|
| Organism |
Sinorhizobium sp. RAC02 [GN: six]
|
|---|
| Gene |
|
|---|
| Compound |
| C01262 | beta-Alanyl-N(pi)-methyl-L-histidine |
| C03492 | D-4'-Phosphopantothenate |
|
|---|
Related
pathway |
| six00250 | Alanine, aspartate and glutamate metabolism |
| six00330 | Arginine and proline metabolism |
| six00770 | Pantothenate and CoA biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|