 | | PATHWAY: stg01110 | |
| Entry |
|
|---|
| Name |
Biosynthesis of secondary metabolites - Streptococcus pyogenes MGAS15252 (serotype M59) |
|---|
| Class |
|
|---|
| Pathway map |
| stg01110 | Biosynthesis of secondary metabolites |
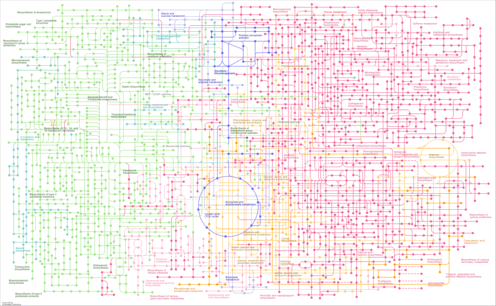
|
|---|
| Module |
| stg_M00022 | Shikimate pathway, phosphoenolpyruvate + erythrose-4P => chorismate [PATH:stg01110] |
|
|---|
| Organism |
Streptococcus pyogenes MGAS15252 (serotype M59) [GN: stg]
|
|---|
Related
pathway |
| stg00053 | Ascorbate and aldarate metabolism |
| stg00250 | Alanine, aspartate and glutamate metabolism |
| stg00260 | Glycine, serine and threonine metabolism |
| stg00270 | Cysteine and methionine metabolism |
| stg00280 | Valine, leucine and isoleucine degradation |
| stg00290 | Valine, leucine and isoleucine biosynthesis |
| stg00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| stg00523 | Polyketide sugar unit biosynthesis |
| stg00525 | Acarbose and validamycin biosynthesis |
| stg00630 | Glyoxylate and dicarboxylate metabolism |
| stg00770 | Pantothenate and CoA biosynthesis |
| stg00900 | Terpenoid backbone biosynthesis |
| stg00999 | Biosynthesis of various plant secondary metabolites |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|