 | | PATHWAY: vca03020 | |
| Entry |
|
|---|
| Name |
RNA polymerase - Vibrio campbellii ATCC BAA-1116 |
|---|
| Class |
Genetic Information Processing; Transcription
|
|---|
| Pathway map |
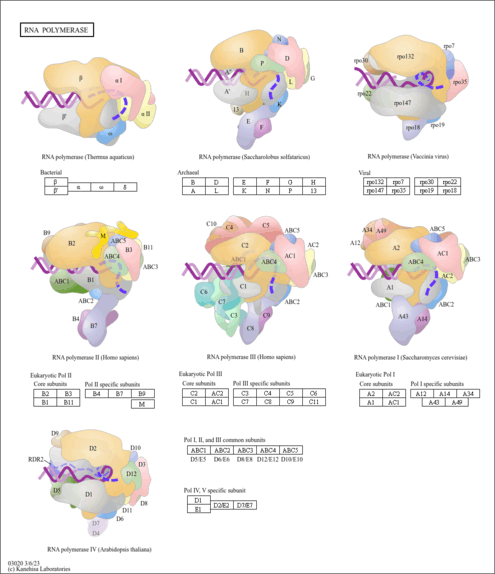
|
|---|
| Other DBs |
|
|---|
| Organism |
Vibrio campbellii ATCC BAA-1116 [GN: vca]
|
|---|
| Gene |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. |
|---|
| Title |
Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. |
|---|
| Title |
Architecture of RNA polymerase II and implications for the transcription mechanism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hirata A, Murakami KS |
|---|
| Title |
Archaeal RNA polymerase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A |
|---|
| Title |
Structure of eukaryotic RNA polymerases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hillen HS, Bartuli J, Grimm C, Dienemann C, Bedenk K, Szalay AA, Fischer U, Cramer P |
|---|
| Title |
Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|