| Entry |
Thumbnail Image |
Name |
Description |
Object |
Legend |
|
map01100
|
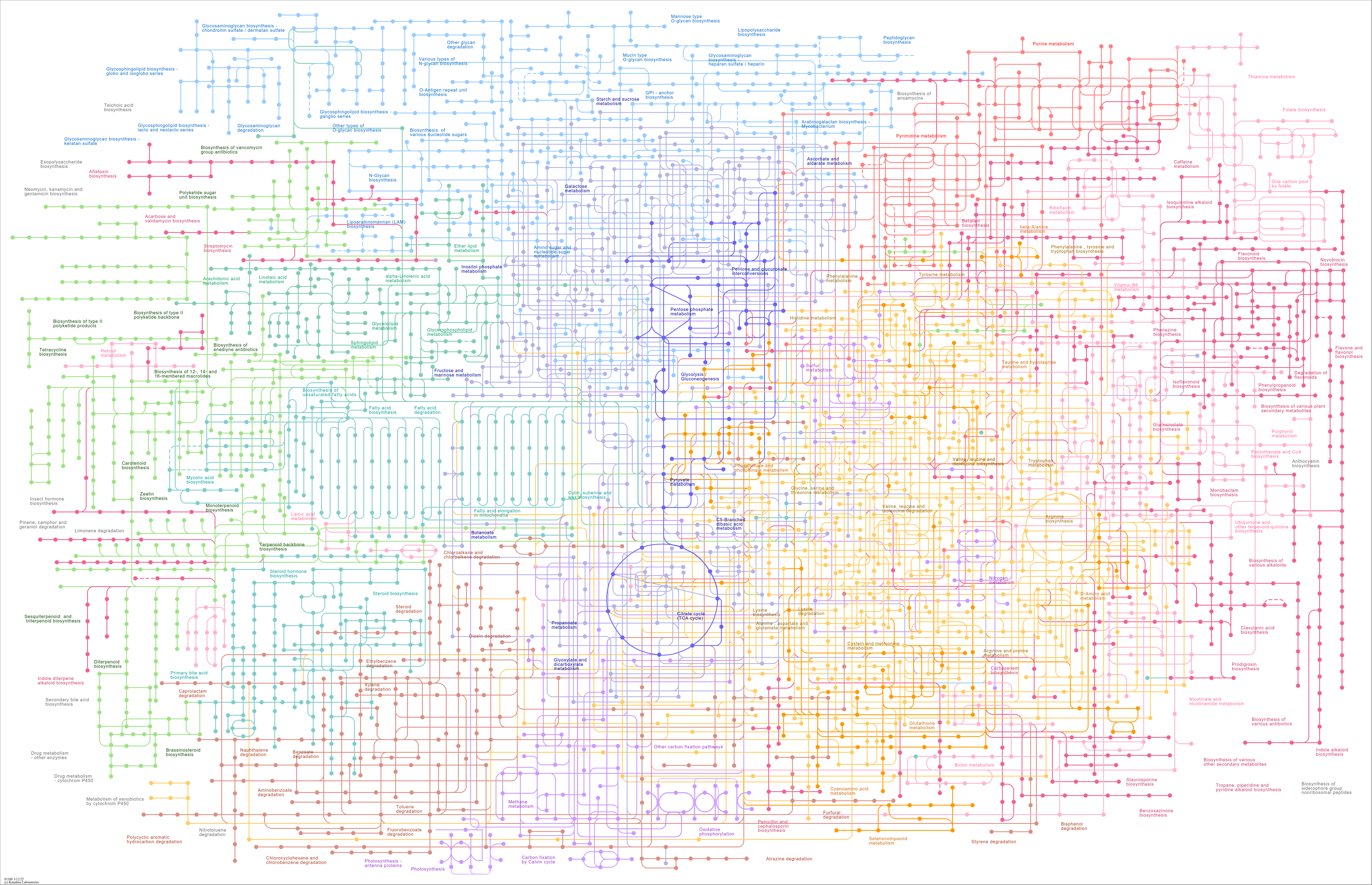
|
Metabolic pathways |
|
...(suyA), K16846 (suyB), 4.4.1.24, R07633 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
Monoterpenoid biosynthesis One carbon pool by folate Flavonoid biosynthesis Starch and sucrose metab... |
|
map01210
|

|
2-Oxocarboxylic acid metabolism |
2-Oxocarboxylic acids, also called 2-oxo acids and alpha-keto acids, are the most elementary set of ... |
...00 (Propanoyl-CoA) C00810 ((R)-Acetoin) K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
8-Methylthiooctyl glucosinolate 2-Oxo-10-methylthio-decanoic acid 7-Methylthioheptyl glucosinolate 2... |
|
map01230
|

|
Biosynthesis of amino acids |
This map presents a modular architecture of the biosynthesis pathways of twenty amino acids, which m... |
..., K12525 (metL), K12526 (lysAC), R00480 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
BIOSYNTHESIS OF AMINO ACIDS Glyceraldehyde-3P Glycerone-P Glycerate-3P Phosphoenolpyruvate Pyruva... |
|
map00270
|
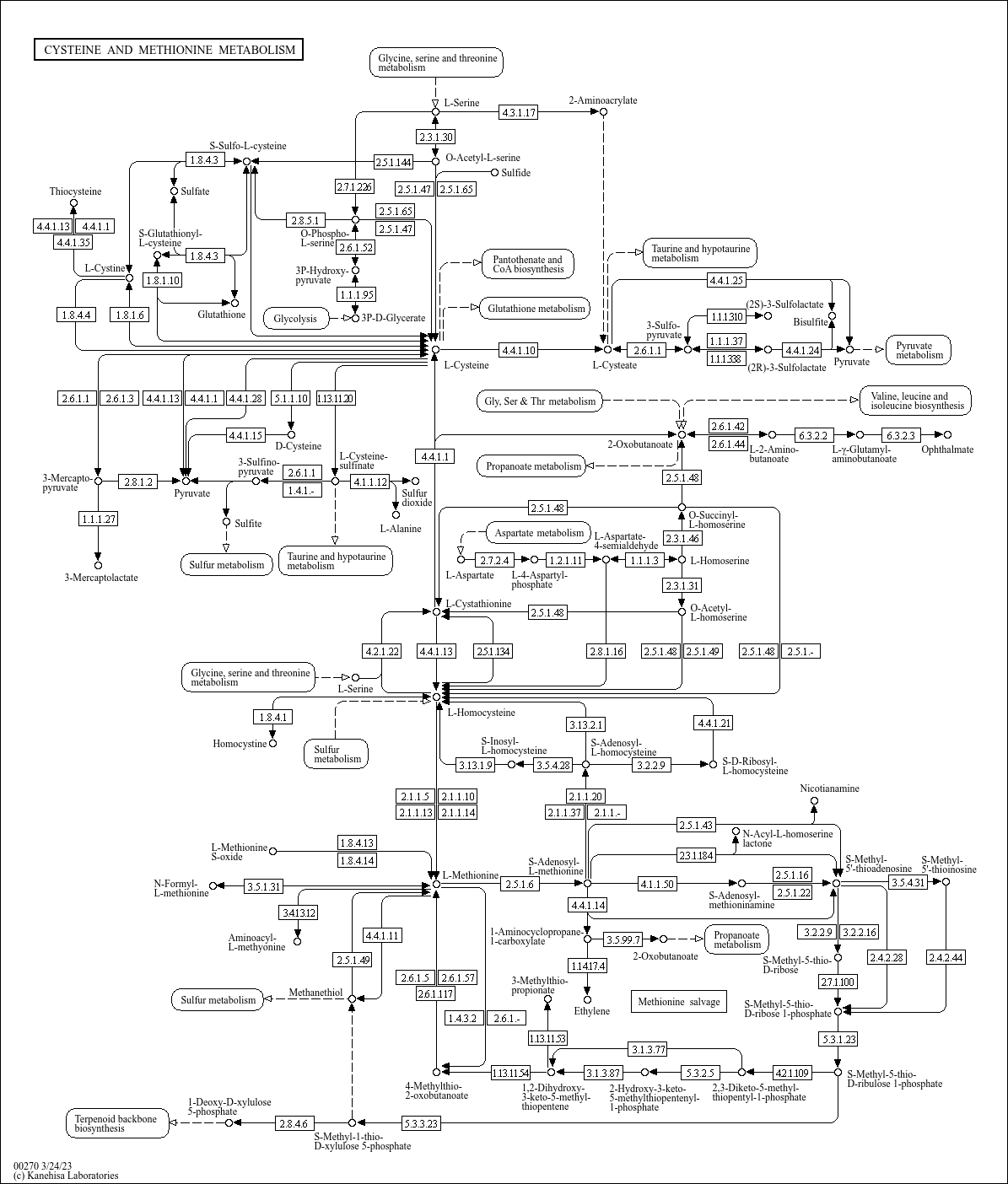
|
Cysteine and methionine metabolism |
Cysteine and methionine are sulfur-containing amino acids. Cysteine is synthesized from serine throu... |
... R08699 K10150 (cysO), 2.5.1.65, R07274 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
Glycolysis 2.5.1.- Ethylene 1-Aminocyclopropane-1-carboxylate 1.14.17.4 4.4.1.14 2.6.1.117 3-Methyl... |
|
map01110
|
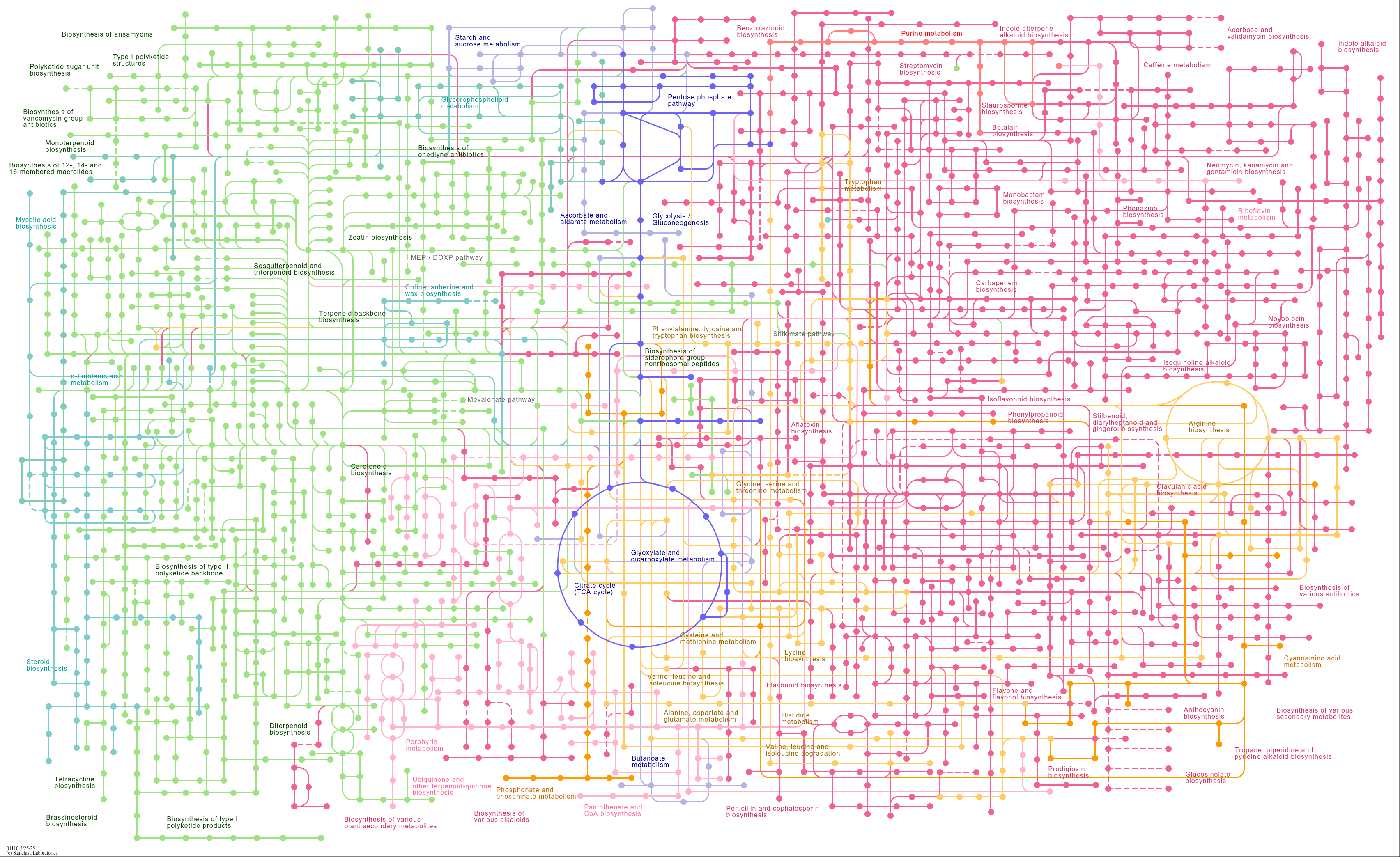
|
Biosynthesis of secondary metabolites |
|
...R1), 1.3.1.12, 1.3.1.13, R01728, R01730 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
Starch and sucrose metabolism Benzoxazinoid biosynthesis Indole diterpene alkaloid biosynthesis Phos... |
|
map01200
|
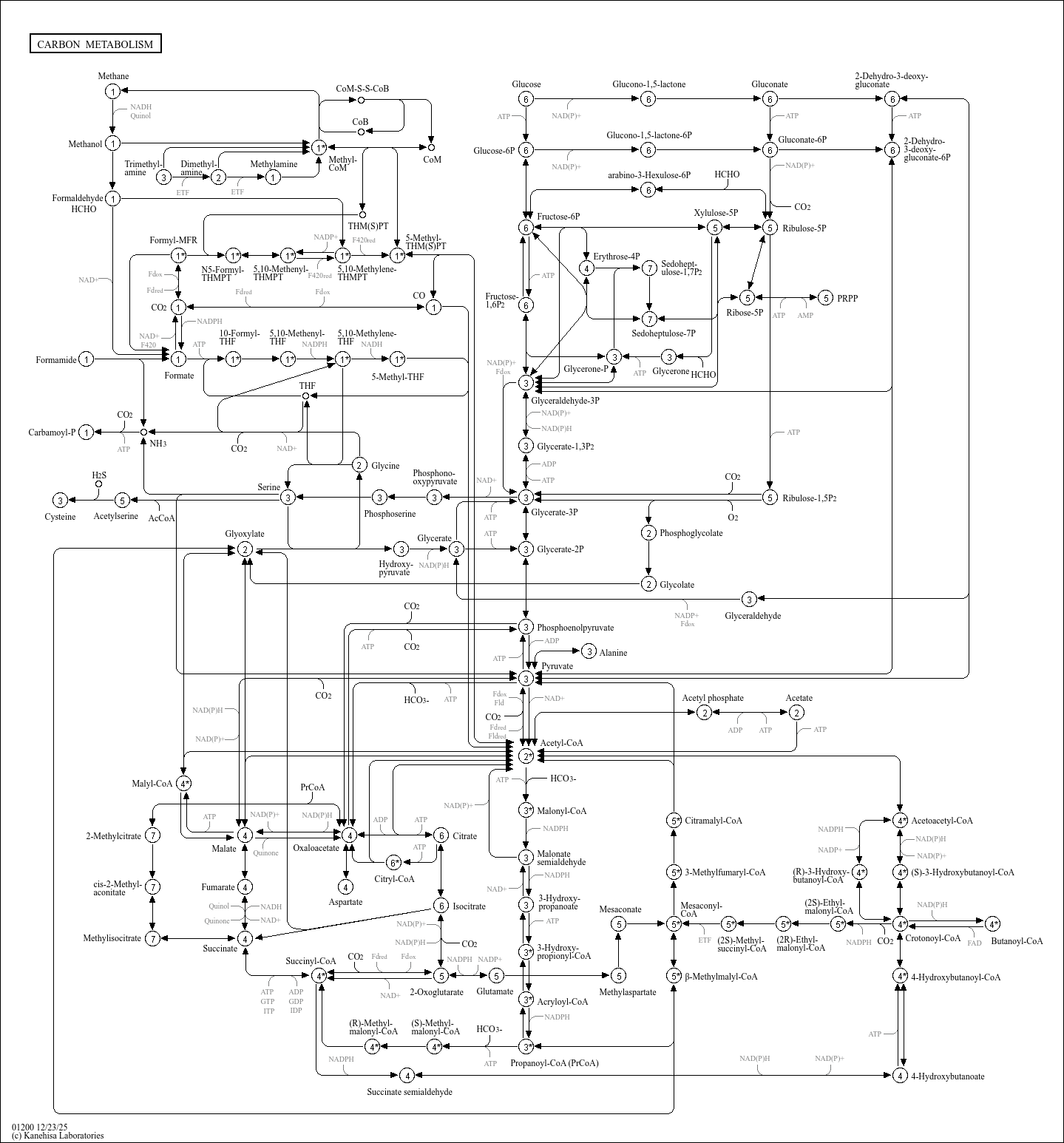
|
Carbon metabolism |
Carbon metabolism is the most basic aspect of life. This map presents an overall view of central car... |
...5, R00706 K00140 (mmsA), R00705, R00706 K14454 (GOT1), K14455 (GOT2), R00355 K14454 (GOT1), K14455 (... |
HCO HCHO HCHO HCO HCO AcCoA CARBON METABOLISM Fructose-6P Ribulose-1,5P Glycerate-3P THF Glucose Gl... |
|
map00400
|

|
Phenylalanine, tyrosine and tryptophan biosynthesis |
|
...(tyrB), K00838 (ARO8), 2.6.1.57, R00734 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
Shikimate pathway 2.5.1.54 Phenylpropanoid biosynthesis Phenylpropanoid biosynthesis Biosynthesis of... |
|
map00710
|
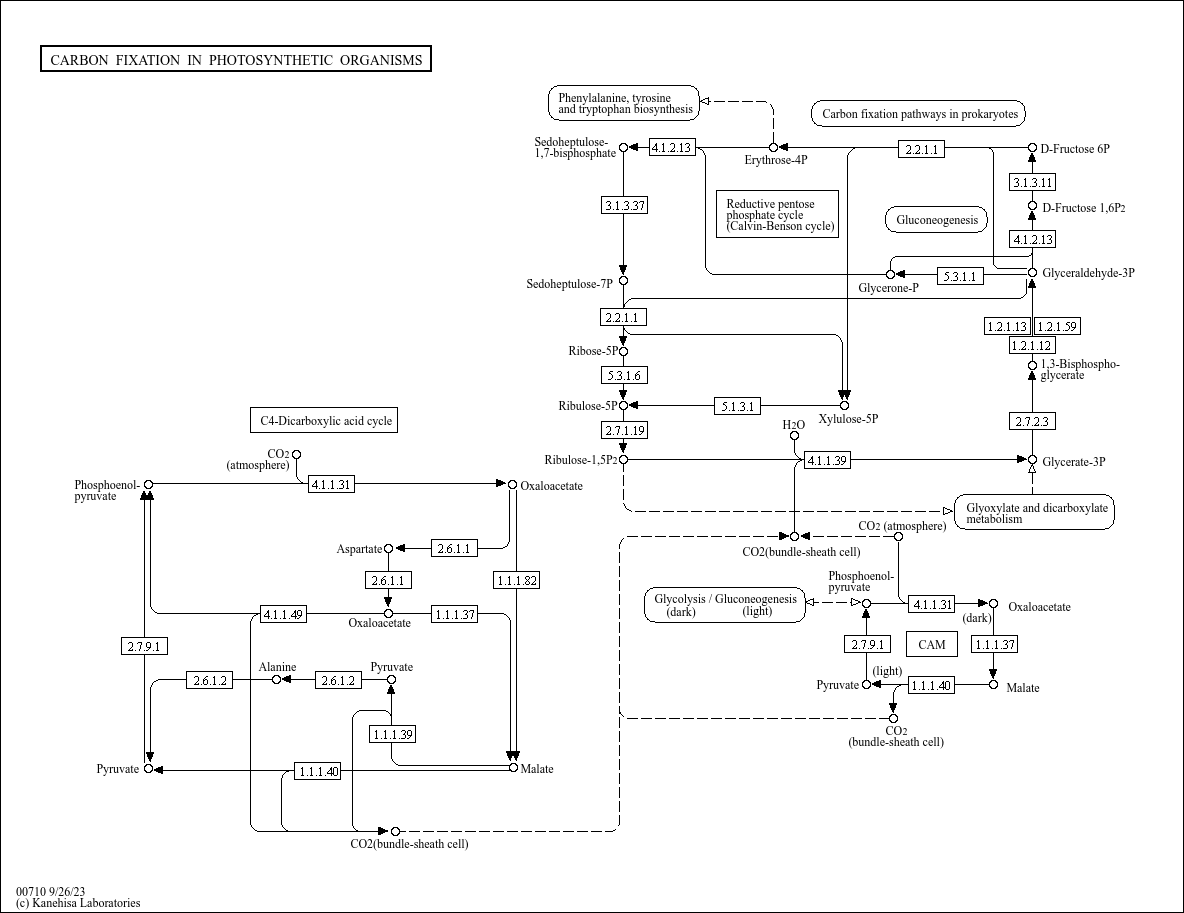
|
Carbon fixation in photosynthetic organisms |
|
...R01063 K00134 (GAPDH), 1.2.1.12, R01061 K14454 (GOT1), 2.6.1.1, R00355 K14454 (GOT1), K14455 (GOT2),... |
(atmosphere) CARBON FIXATION IN PHOTOSYNTHETIC ORGANISMS 4.1.2.13 3.1.3.37 2.2.1.1 5.3.1.6 2.7.1... |
|
map00220
|
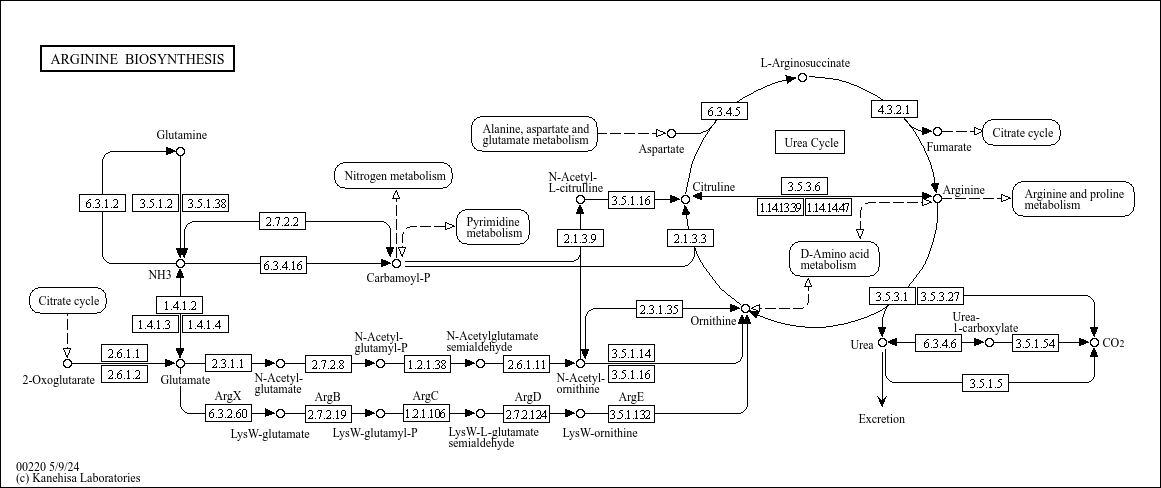
|
Arginine biosynthesis |
|
...R00256 K00491 (nos), 1.14.14.47, R11711 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
L-Arginosuccinate Citruline Arginine Ornithine Carbamoyl-P Aspartate Fumarate NH3 N-Acetyl-glutamate... |
|
map00250
|
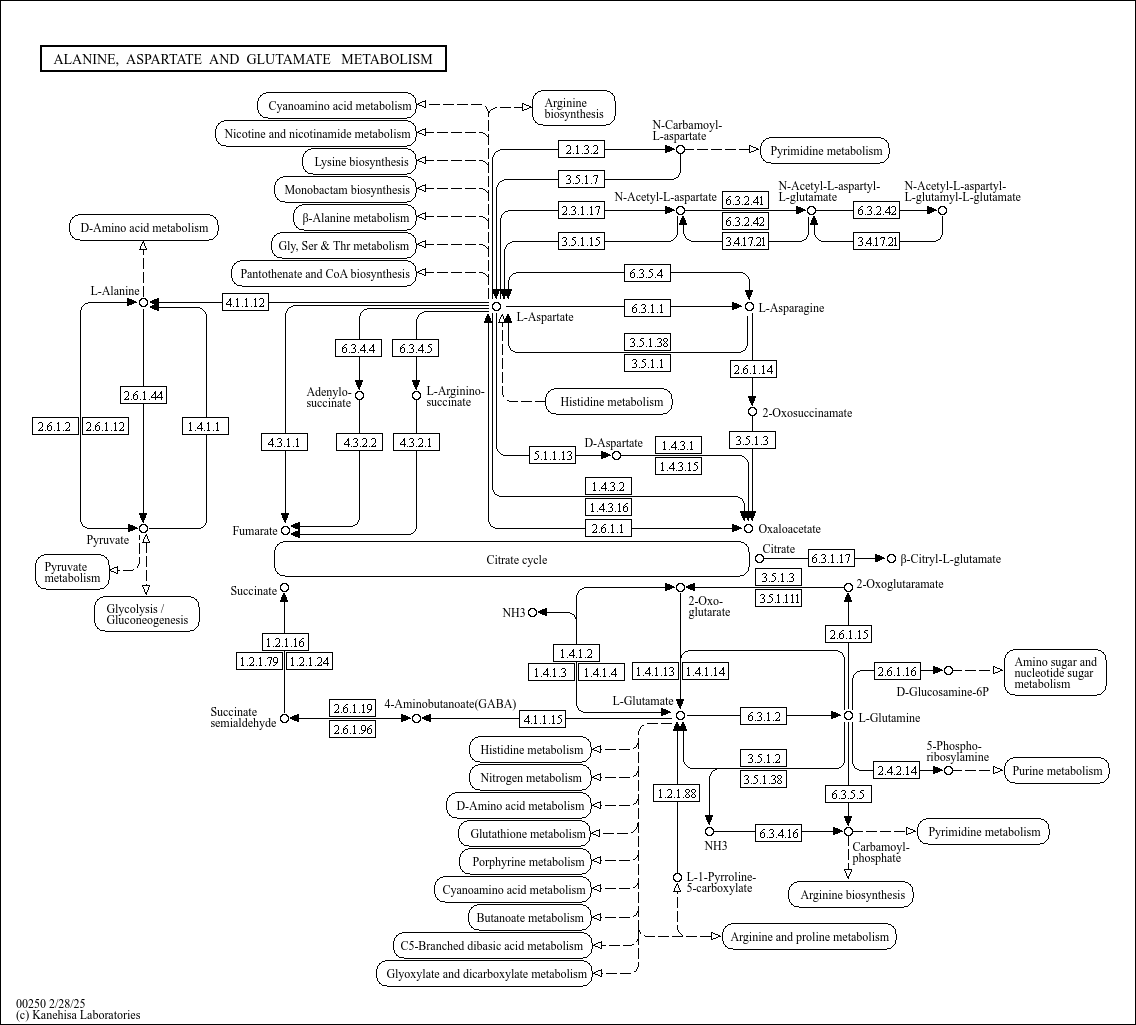
|
Alanine, aspartate and glutamate metabolism |
|
... R00357 K00278 (nadB), 1.4.3.16, R00357 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
Fumarate Succinate Oxaloacetate 2-Oxo-glutarate Citrate cycle L-Alanine L-Aspartate L-Asparagine D-A... |
|
map00330
|
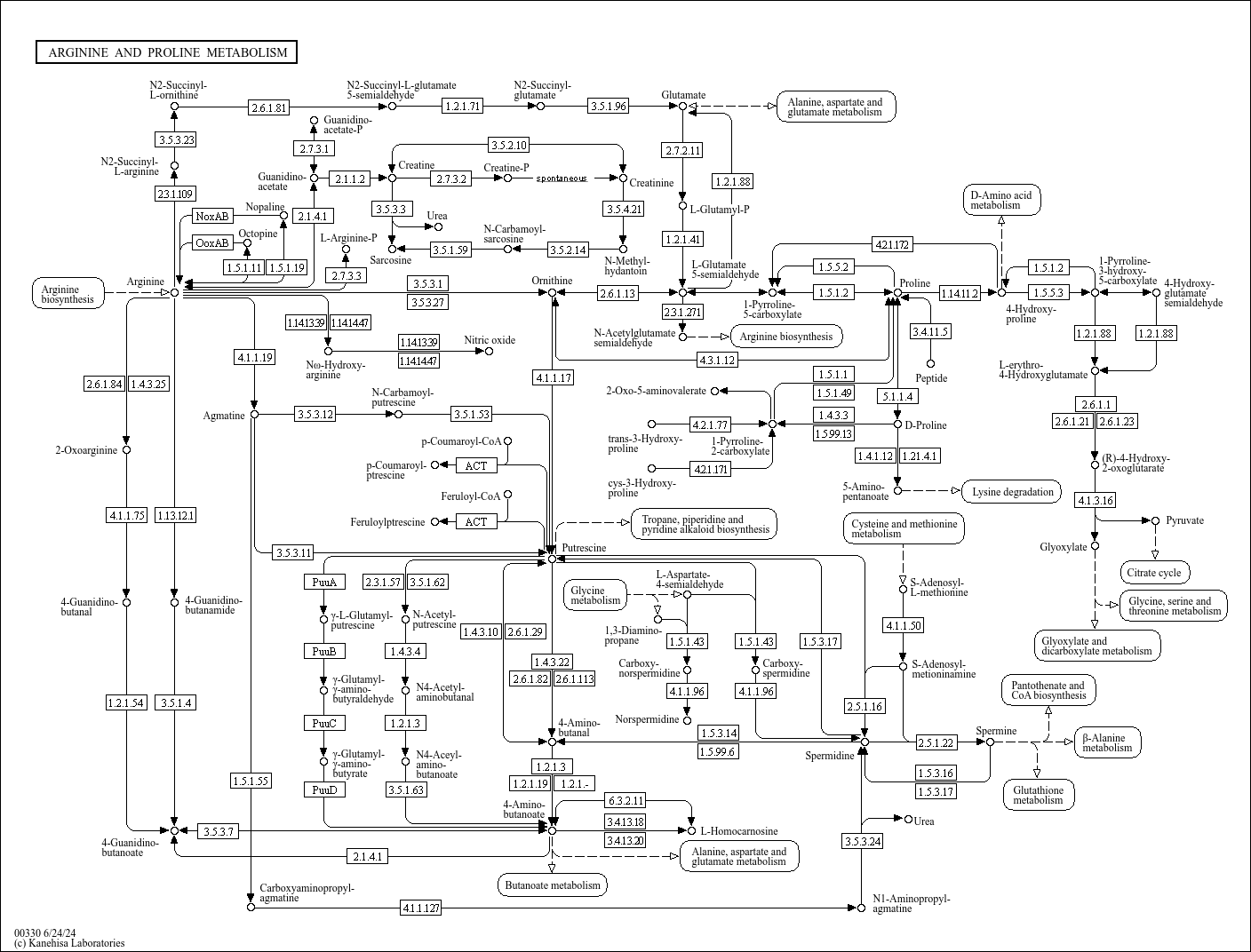
|
Arginine and proline metabolism |
|
...R12085 K23121 (pflD), 4.2.1.172, R11819 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
1.5.1.11 Octopine OoxAB ARGININE AND PROLINE METABOLISM Polyamine Pathway ADC pathway ADH pathway... |
|
map00350
|

|
Tyrosine metabolism |
|
... R03181 K00451 (HGD), 1.13.11.5, R02519 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
TYROSINE METABOLISM 1.14.18.1 4.1.99.2 1.10.3.1 Phenol 1.11.1.8 1.11.1.8 1.11.1.8 1.11.1.8 3-Iodo-L... |
|
map00360
|

|
Phenylalanine metabolism |
|
...5 (TAT), K00838 (ARO8), 2.6.1.5, R00694 K14454 (GOT1), K14455 (GOT2), K00811 (ASP5), K00812 (aspB), ... |
4.1.3.39 4.3.1.25 Tropane, piperidine and pyridine alkaloid biosynthesis 1.2.1.5 6.2.1.30 2.3.1.14 4... |
|
map00950
|

|
Isoquinoline alkaloid biosynthesis |
Isoquinoline alkaloids are tyrosine-derived plant alkaloids with an isoquinoline skeleton. Among the... |
...(TYDC), 4.1.1.25, R00736 K14455 (GOT2), K14454 (GOT1), K00811 (ASP5), K00812 (aspB), K00813 (aspC), ... |
1.1.1.218 2.3.1.150 2.6.1.5 1.5.1.27 1.14.19.67 1.14.19.65 1.14.19.64 2.1.1.122 1.14.14.97 1.14.14.9... |
|
map00960
|

|
Tropane, piperidine and pyridine alkaloid biosynthesis |
|
...(ARO8), 2.6.1.57, R00694 K14455 (GOT2), K14454 (GOT1), K00811 (ASP5), K00812 (aspB), K00813 (aspC), ... |
Local analgesic Cadaverine Phenylalanine,tyrosine and tryptophan biosynthesis Phenylalanine metaboli... |
|
map01120
|

|
Microbial metabolism in diverse environments |
|
..., R05581 K07539 (oah), 3.7.1.21, R05594 K14454 (GOT1), K14455 (GOT2), 2.6.1.1, R00355 K01595 (ppc), ... |
Chlorocyclohexane and chlorobenzene degradation Xylene degradation Aminobenzoate degradation Naphtha... |