| Entry |
|
|---|
| Name |
Furfural degradation - Neorhizobium petrolearium |
|---|
| Class |
Metabolism; Xenobiotics biodegradation and metabolism
|
|---|
| Pathway map |
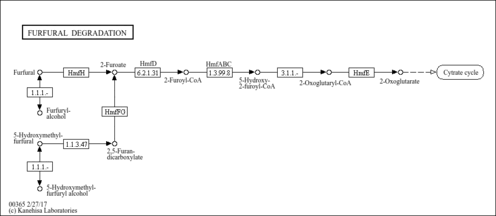
|
|---|
| Organism |
Neorhizobium petrolearium [GN: npm]
|
|---|
| Gene |
|
|---|
| Compound |
| C03724 | S-(5-Hydroxy-2-furoyl)-CoA |
| C11101 | 5-Hydroxymethyl-2-furaldehyde |
| C20443 | 5-Hydroxymethylfurfuryl alcohol |
|
|---|
| Reference |
|
|---|
| Authors |
Wierckx N, Koopman F, Ruijssenaars HJ, de Winde JH |
|---|
| Title |
Microbial degradation of furanic compounds: biochemistry, genetics, and impact. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ |
|---|
| Title |
Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|