| Entry |
Thumbnail Image |
Name |
Description |
Object |
Legend |
|
map00627
|
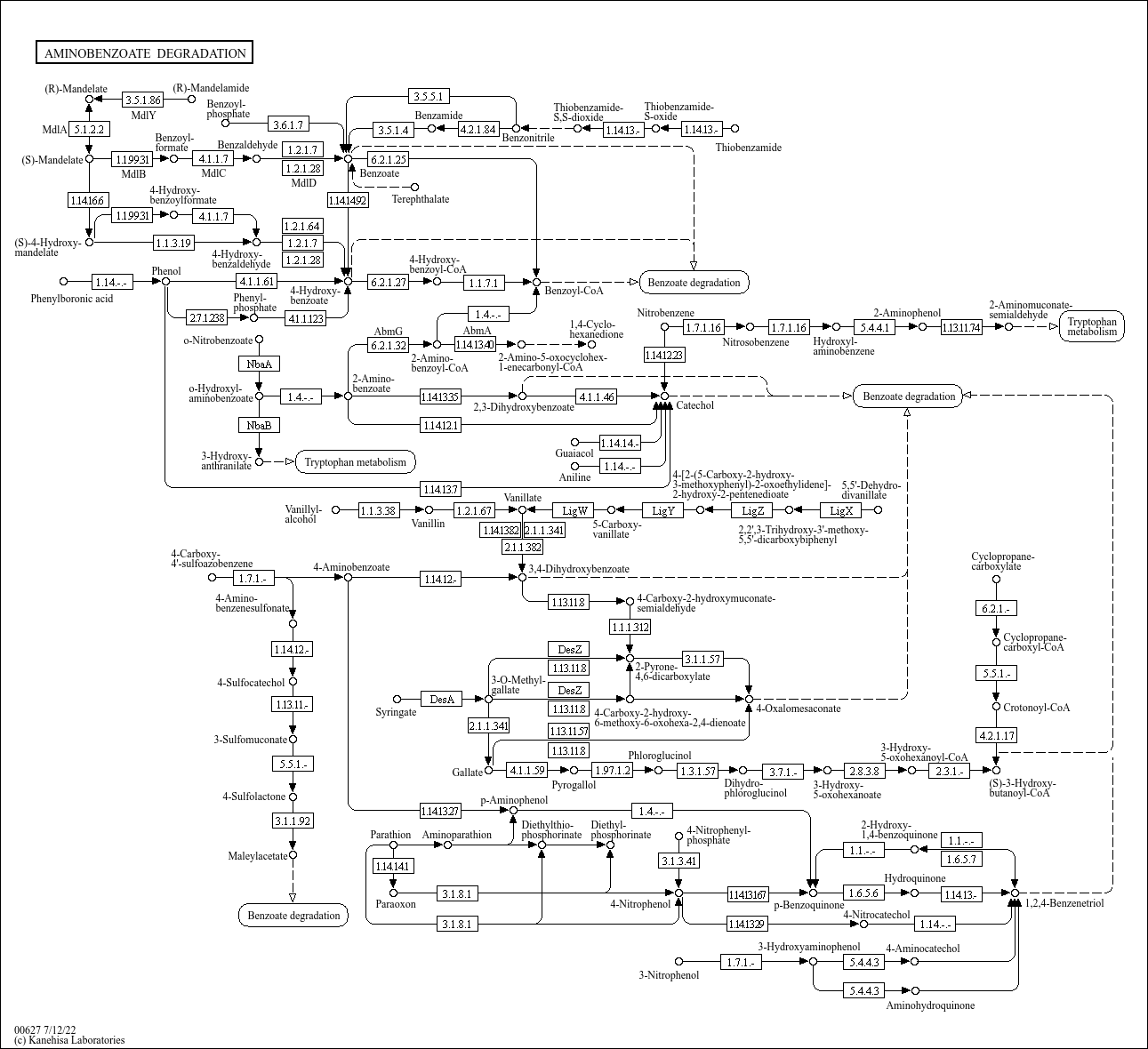
|
Aminobenzoate degradation |
|
C00568 (4-Aminobenzoate) C00230 (3,4-Dihydroxybenzoate) C07103 (2-Hydroxy-1,4-benzoquinone) C03360 (... |
AMINOBENZOATE DEGRADATION 4-Aminobenzoate 1.14.12.- 1.7.1.- 3,4-Dihydroxybenzoate 3.1.8.1 2-Hydrox... |
|
map00362
|

|
Benzoate degradation |
|
...1-ene-1,2,4-tricarboxylate) C00587 (3-Hydroxybenzoate) C00628 (2,5-Dihydroxybenzoate) C00156 (4-Hydr... |
3-Hydroxy-pimeloyl-CoA BENZOATE DEGRADATION 3-Carboxy-2,5-dihydro-5-oxofuran-2-acetate 1.14.13.33... |
|
map00364
|
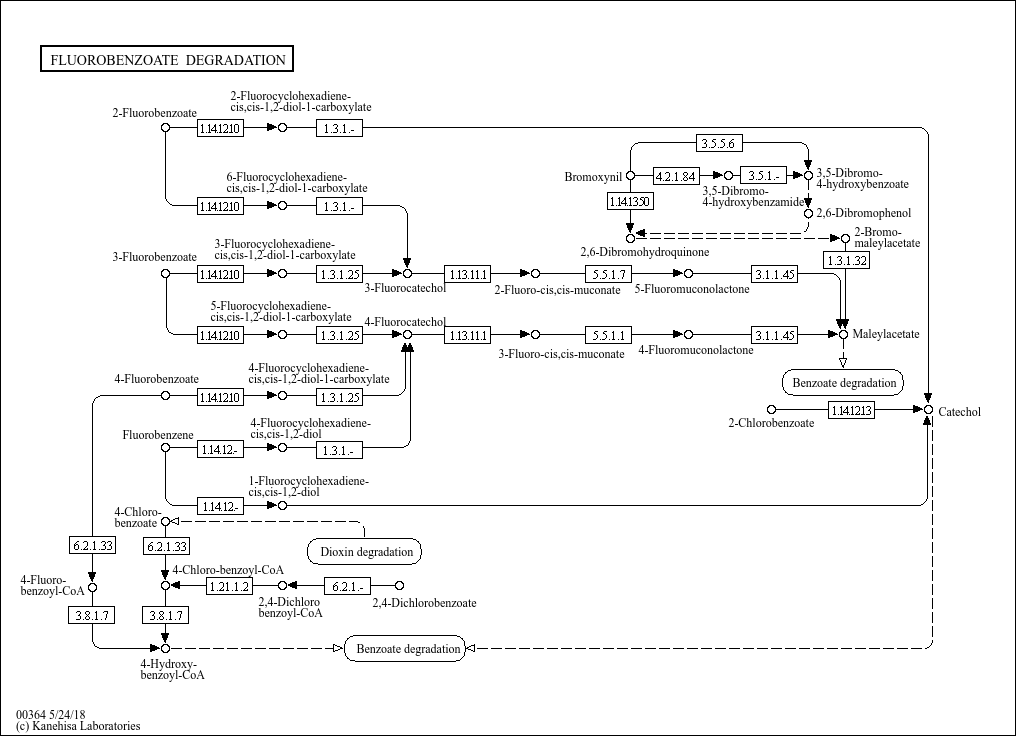
|
Fluorobenzoate degradation |
|
C02364 (3-Fluorobenzoate) C16474 (3-Fluoro-cis,cis-muconate) C00090 (Catechol) C16484 (1-Fluorocyclo... |
3-Fluorobenzoate 1.14.12.- 1.14.12.- 3.1.1.45 3.1.1.45 3-Fluoro-cis,cis-muconate Catechol 1-Fluorocy... |
|
map00640
|
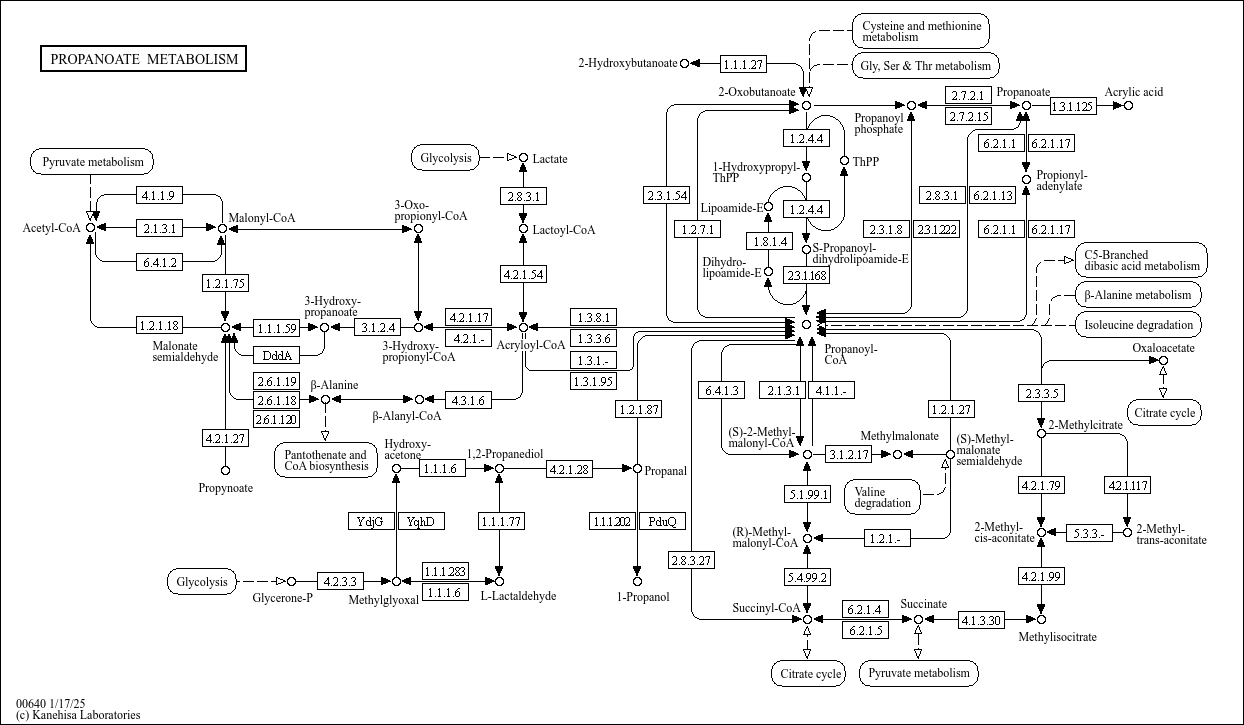
|
Propanoate metabolism |
|
C00222 (3-Oxopropanoate) C00804 (Propynoate) C01013 (3-Hydroxypropanoate) C00099 (beta-Alanine) C000... |
PROPANOATE METABOLISM 1.2.1.18 2.1.3.1 6.4.1.2 4.1.1.9 1.2.1.75 2.6.1.18 2.6.1.19 1.1.1.59 4.2.1.27... |
|
map00650
|
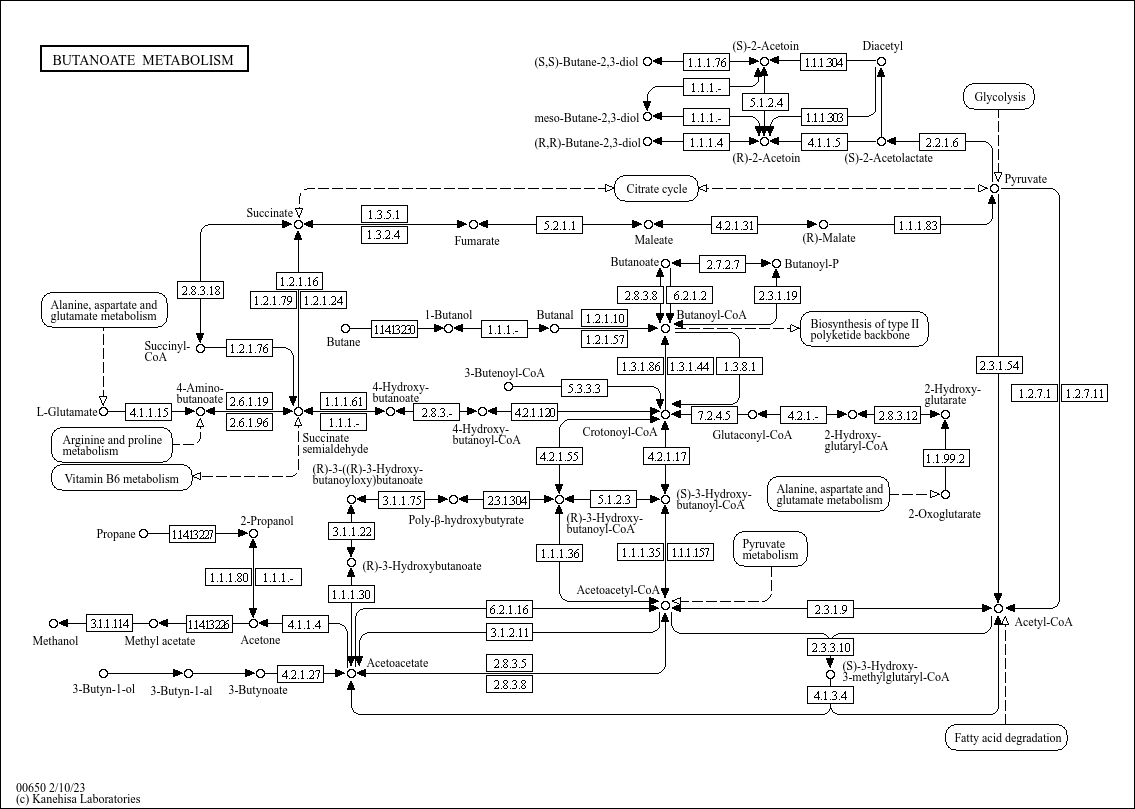
|
Butanoate metabolism |
|
... C00164 (Acetoacetate) C01089 ((R)-3-Hydroxybutanoate) C04546 ((R)-3-((R)-3-Hydroxybutanoyloxy)butan... |
...e metabolism Pyruvate metabolism Glycolysis BUTANOATE METABOLISM Citrate cycle Vitamin B6 metabolis... |
|
map01210
|

|
2-Oxocarboxylic acid metabolism |
...f metabolites that includes pyruvate (2-oxopropanoate), 2-oxobutanoate, oxaloacetate (2-oxosuccinate... |
...) C00049 (L-Aspartate) C03283 (L-2,4-Diaminobutanoate) C00026 (2-Oxoglutarate) C00322 (2-Oxoadipate)... |
...osinolate Oxaloacetate Aspartate 2,4-Diaminobutanoate 2-Oxoglutarate 2-Oxoadipate Glutamate Ornithin... |
|
map00130
|

|
Ubiquinone and other terpenoid-quinone biosynthesis |
...derived from the shikimate pathway; 4-hydroxybenzoate is directly formed from chorismate in bacteria... |
...ene-1-carboxylate) C03657 (1,4-Dihydroxy-2-naphthoate) C06986 (Spirodilactone) C00885 (Isochorismate... |
...ne-1-carboxylate 4.2.99.20 1,4-Dihydroxy-2-naphthoate Spirodilactone Phenylalanine, tyrosine and try... |
|
map04142
|

|
Lysosome |
...-Golgi network. They are packaged into clathrin-coated vesicles and are transported to late endosome... |
C00002 (ATP) C00008 (ADP) C00159 (D-Mannose) C00159 (D-Mannose) C00275 (D-Mannose 6-phosphate) C0027... |
...ALP70 sortilin CLN7 HGSNAT Golgi body clathrin coat mannose clathrin M6P receptor from MPR NAGPA la... |
|
map04062
|
|
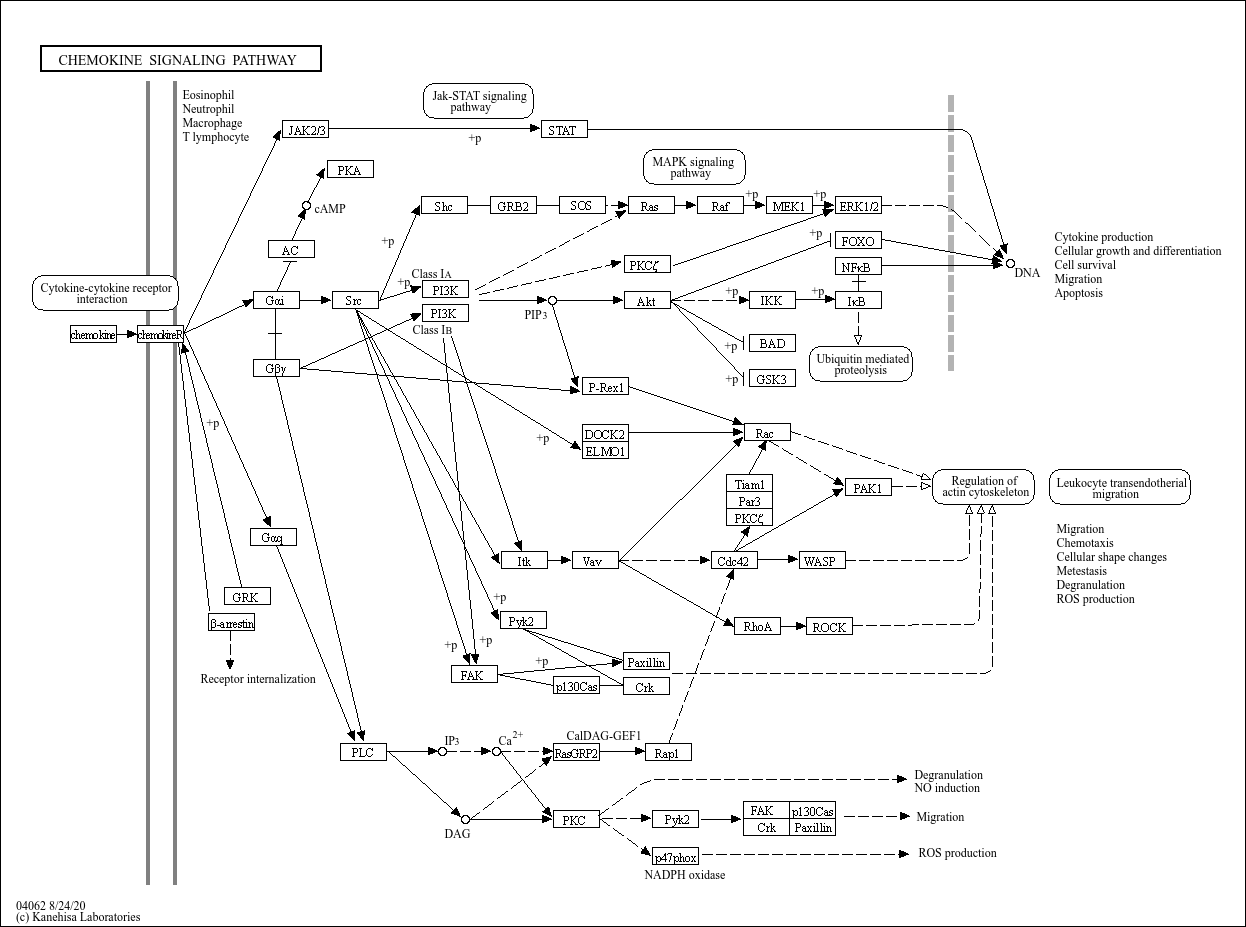
Chemokine signaling pathway |
...on upon foreign insult. Chemokines are small chemoattractant peptides that provide directional cues ... |
C00165 (Diacylglycerol) C01245 (D-myo-Inositol 1,4,5-trisphosphate) C00076 (Calcium cation) C00575 (... |
Migration Apoptosis Degranulation Cellular shape changes Cell survival chemokineR chemokine JAK2/3 ... |
|
map04613
|
|
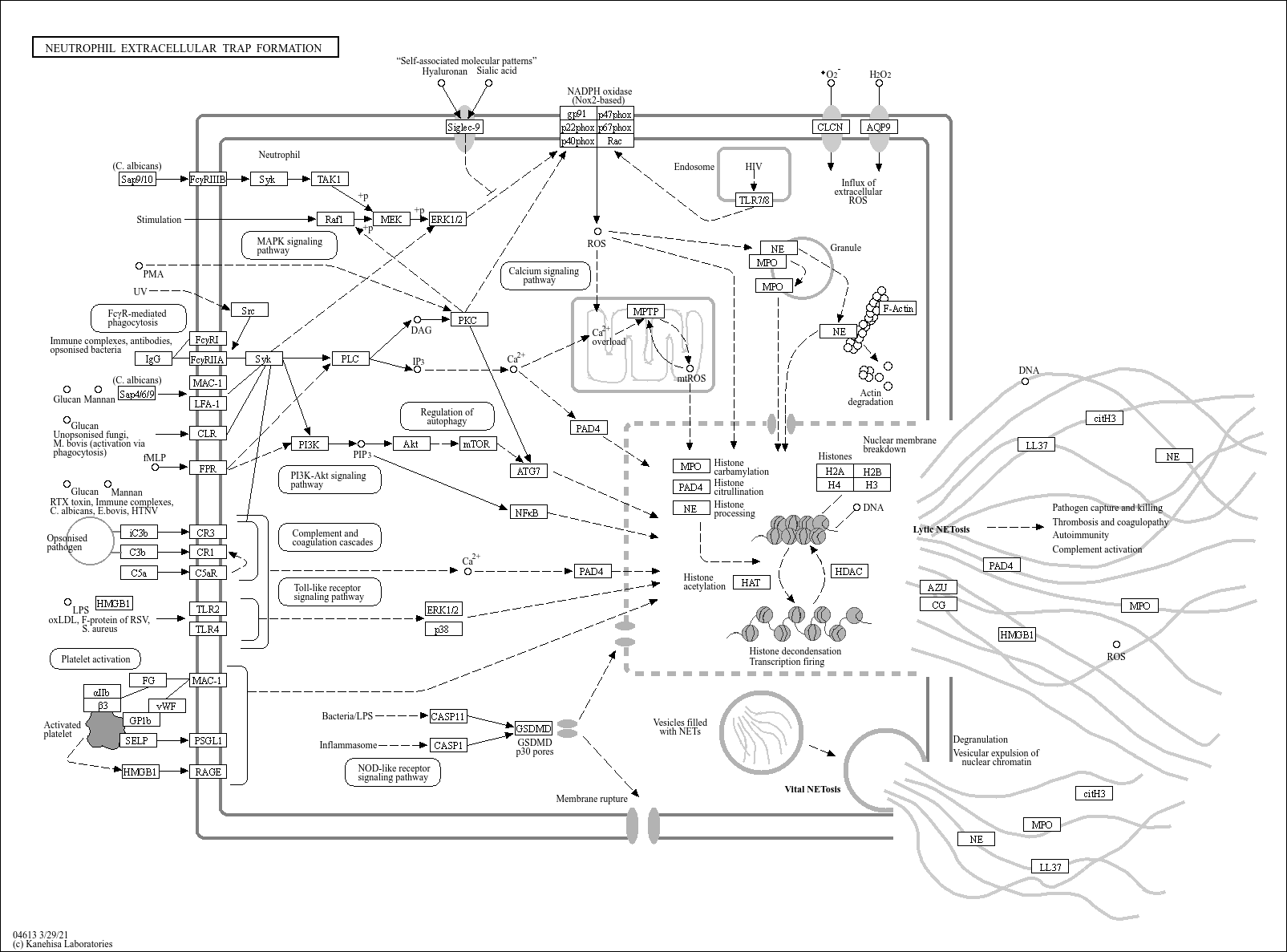
Neutrophil extracellular trap formation |
... extracellular structures composed of chromatin coated with histones, proteases and granular and cyt... |
C05981 (Phosphatidylinositol-3,4,5-trisphosphate) C01245 (D-myo-Inositol 1,4,5-trisphosphate) C00165... |
Neutrophil FcγRIIA FcγRI Src Syk IgG PI3K PIP PLC DAG PKC Raf1 MEK ERK1/2 Akt NOD-like receptor sign... |
|
map01100
|
|
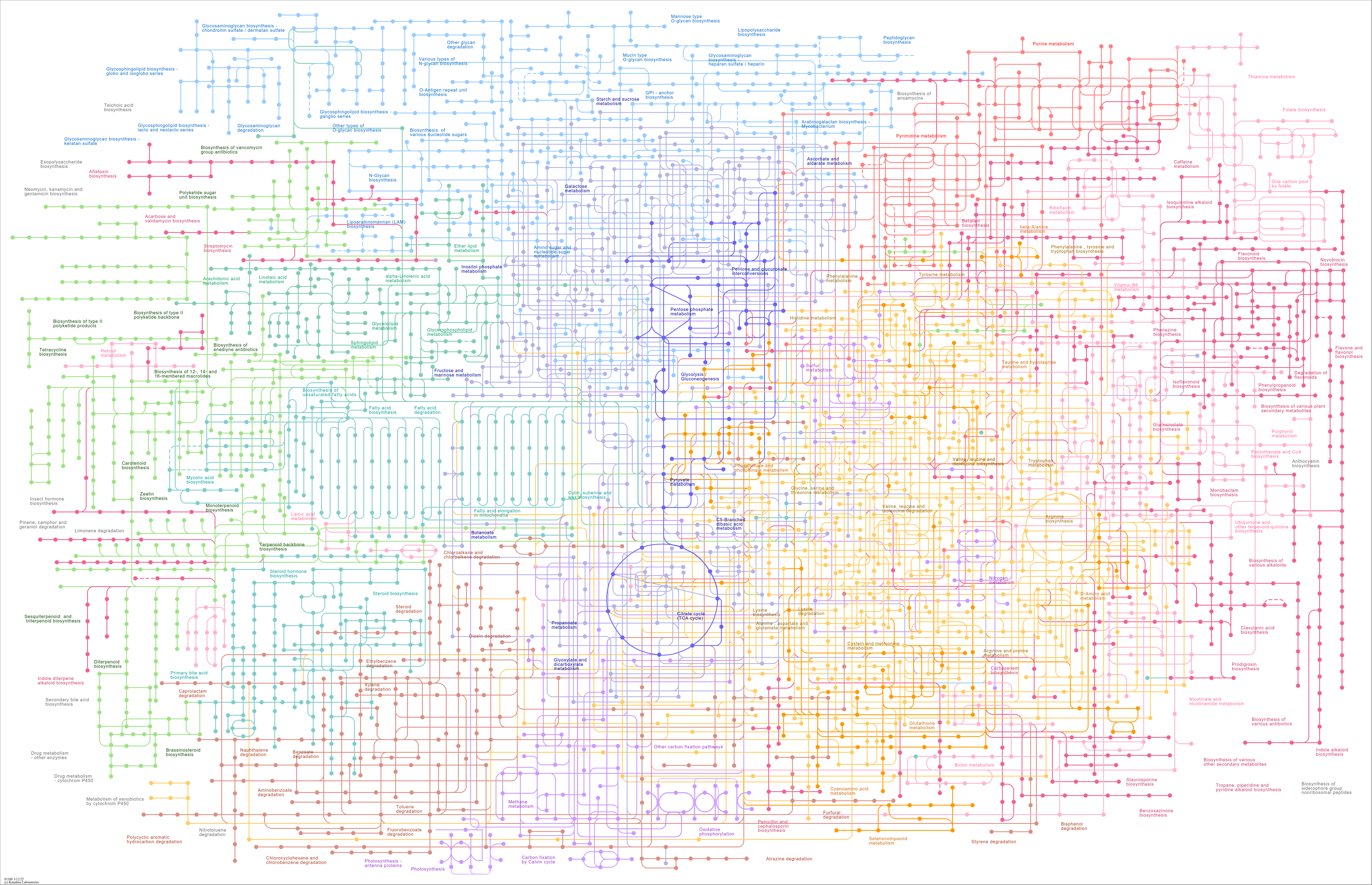
Metabolic pathways |
|
...oA) C06582 (2-Hydroxy-6-oxo-7-methylocta-2,4-dienoate) C00261 (Benzaldehyde) C01841 (Pentalenene) C2... |
...erpenoid and triterpenoid biosynthesis Aminobenzoate degradation Atrazine degradation Fatty acid de... |
|
map01120
|

|
Microbial metabolism in diverse environments |
|
...exanol) C11249 (Cyclohexane) C02378 (6-Aminohexanoate) C06593 (epsilon-Caprolactam) C06105 (Cyclohex... |
...obenzene degradation Xylene degradation Aminobenzoate degradation Naphthalene degradation Dioxin deg... |
|
map01220
|

|
Degradation of aromatic compounds |
Microorganisms are known to be capable of degrading diverse chemical substances including man-made c... |
...enzaldehyde) C00556 (Benzyl alcohol) C00180 (Benzoate) C06321 ((1R,6S)-1,6-Dihydroxycyclohexa-2,4-di... |
...OMPOUNDS Toluene Benzaldehyde Benzyl alcohol Benzoate Catechol 3-Fluorobenzoate 4-Fluorocatechol 4-F... |
|
map01110
|
|
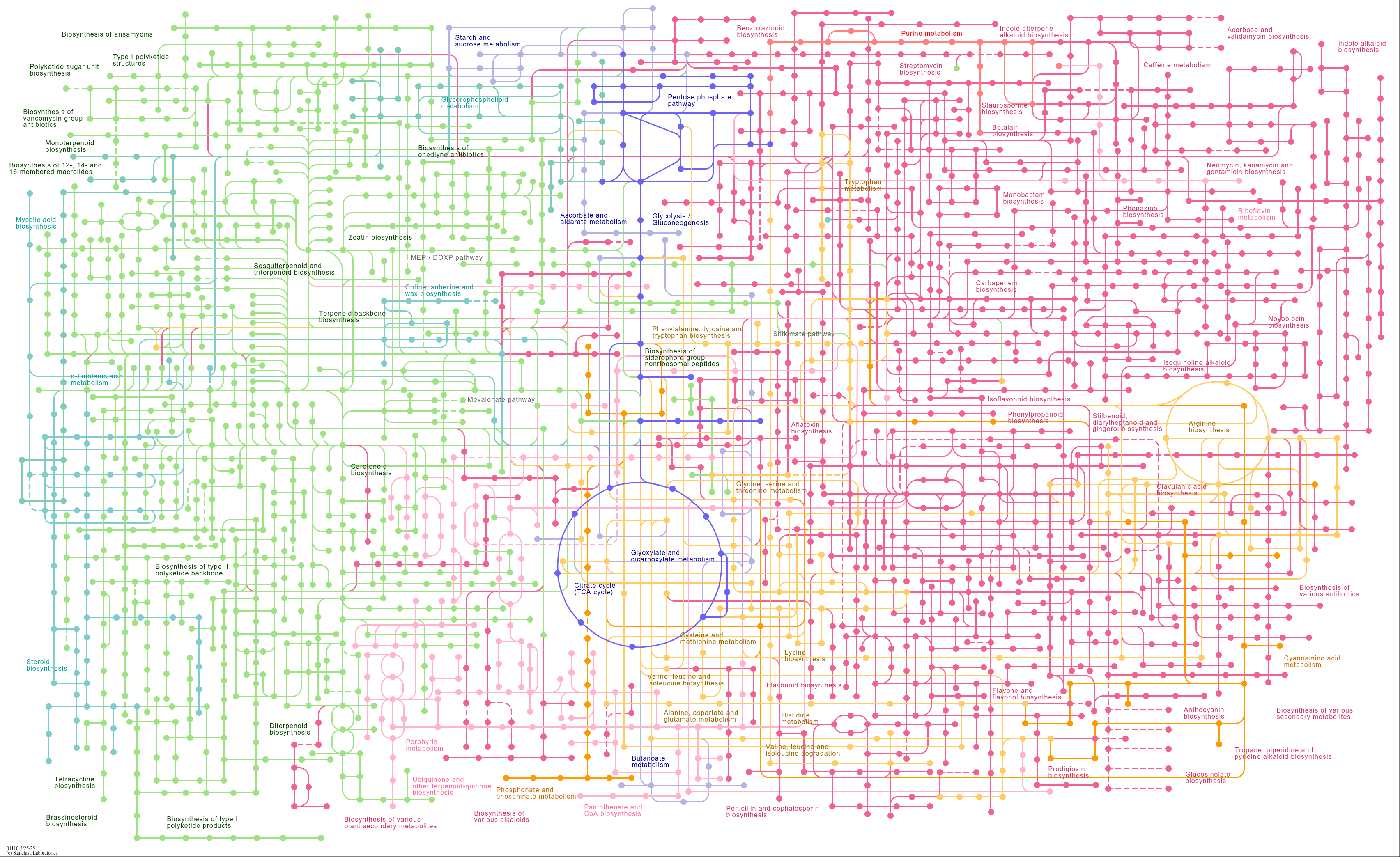
Biosynthesis of secondary metabolites |
|
...oA) C04171 ((2S,3S)-2,3-Dihydro-2,3-dihydroxybenzoate) C00196 (2,3-Dihydroxybenzoate) C05821 (Entero... |
...s Penicillin and cephalosporin biosynthesis Butanoate metabolism Mevalonate pathway Biosynthesis of ... |
|
map01240
|
|
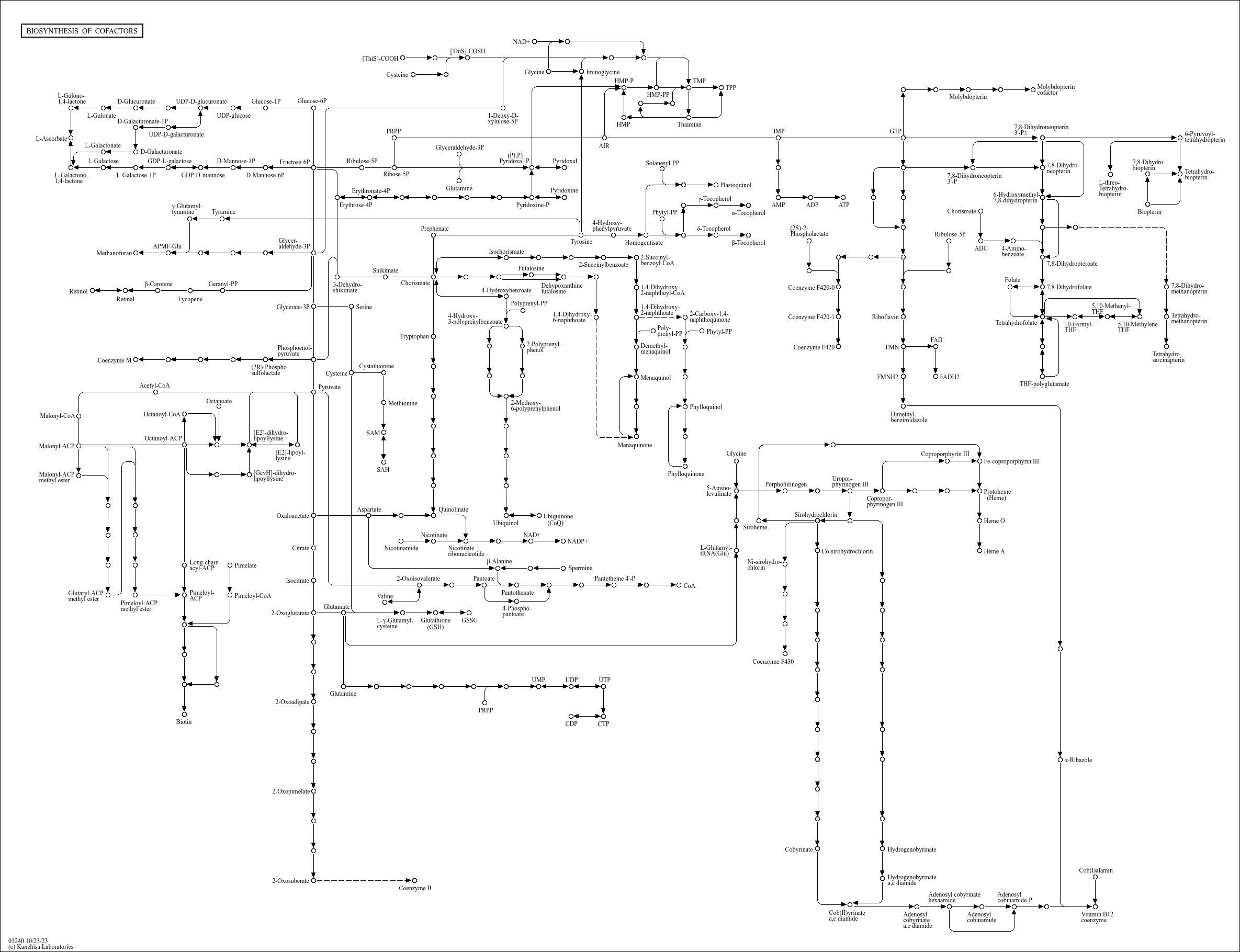
Biosynthesis of cofactors |
|
...oic acid) C00183 (L-Valine) C00966 (2-Dehydropantoate) C00864 (Pantothenate) C00522 ((R)-Pantoate) C... |
...+ NADP+ 2-Oxoisovalerate Valine Pantothenate Pantoate β-Alanine Pantetheine 4'-P CoA Cysteine 7,8-Di... |
|
map00621
|
|
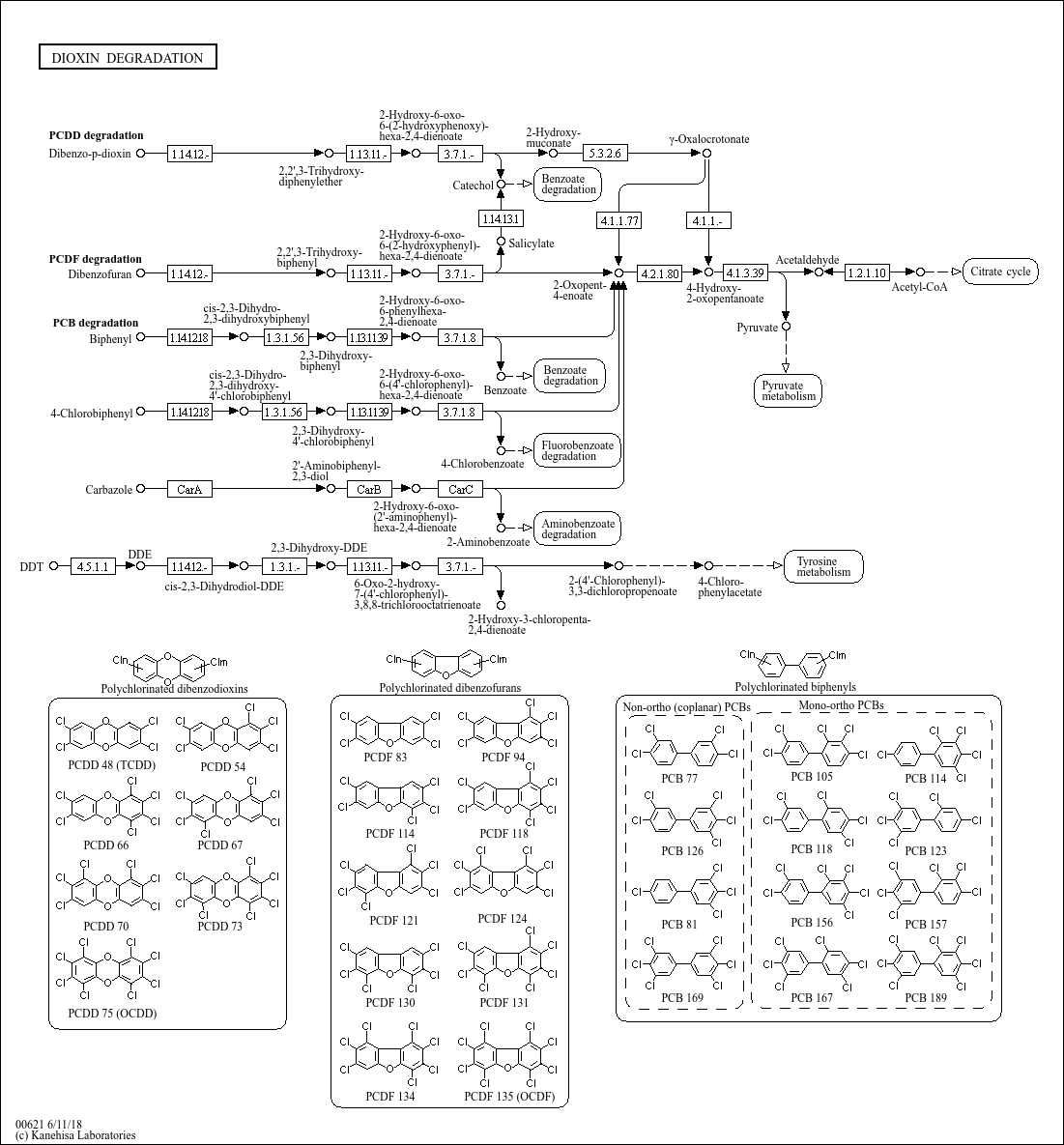
Dioxin degradation |
Dioxins and dioxin-like compounds, including PCDDs, PCDFs and PCBs, are persistent organic pollutant... |
...hydroxybiphenyl) C00805 (Salicylate) C00180 (Benzoate) C06584 (4-Chlorobiphenyl) C06585 (cis-2,3-Dih... |
...late 1.14.12.- 1.14.12.18 1.14.12.- 3.7.1.8 Benzoate 4-Chlorobiphenyl cis-2,3-Dihydro-2,3-dihydroxy... |
|
map01230
|

|
Biosynthesis of amino acids |
This map presents a modular architecture of the biosynthesis pathways of twenty amino acids, which m... |
C04390 (N6-Acetyl-LL-2,6-diaminoheptanedioate) C04002 ((Z)-But-1-ene-1,2,4-tricarboxylate) C00118 (D... |
...bulose-5P Ribose-5P PRPP Erythrose-4P 2-Oxo-butanoate 2-Oxoisovalerate Valine 3-Methyl-2-oxopentanoa... |
|
map00624
|
|
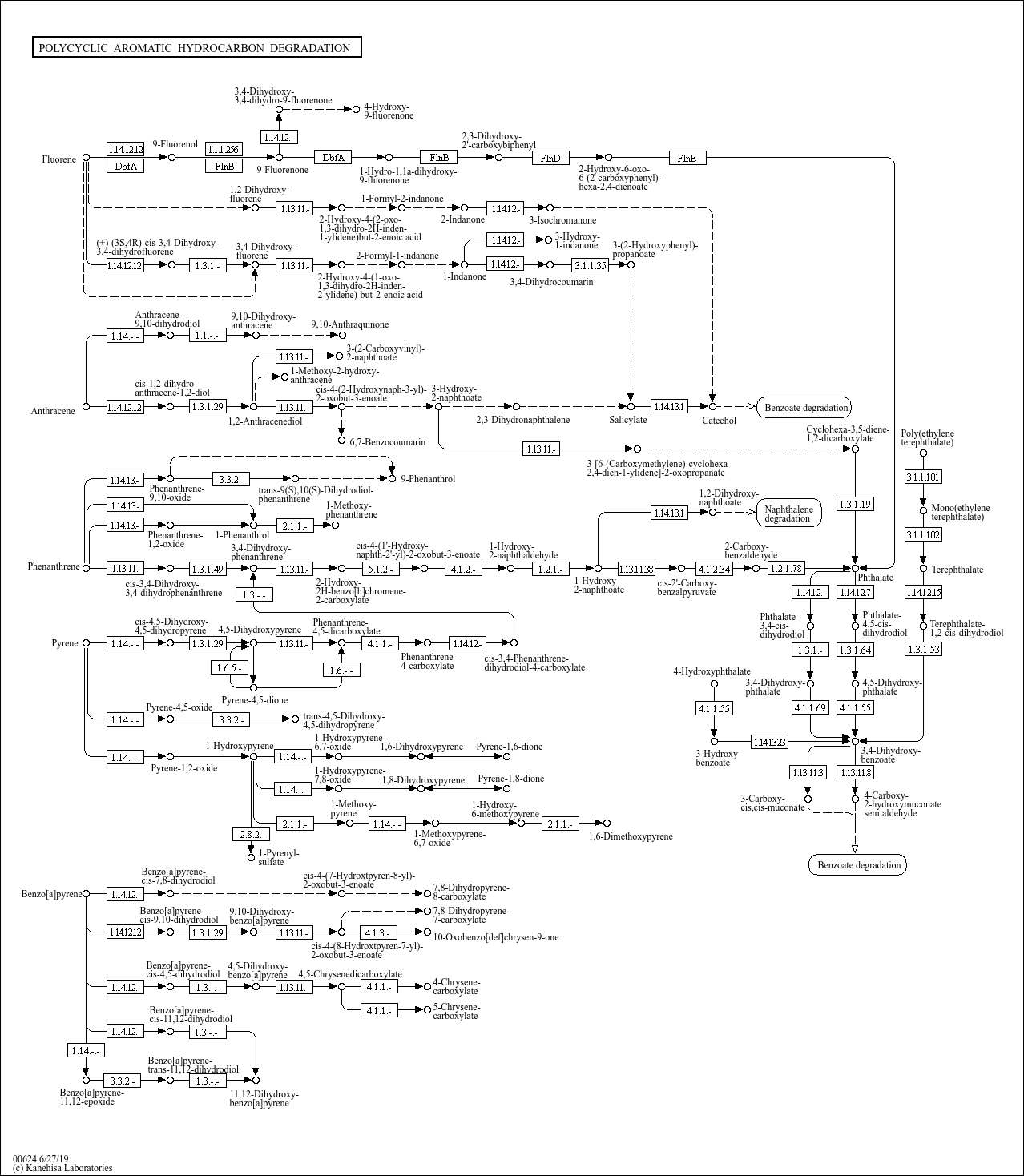
Polycyclic aromatic hydrocarbon degradation |
|
...e) C16149 ((3Z)-4-(2-Carboxyphenyl)-2-oxobut-3-enoate) C11425 (2-Hydroxy-2H-benzo[h]chromene-2-carbo... |
...ate cis-4-(1'-Hydroxy-naphth-2'-yl)-2-oxobut-3-enoate 3,4-Dihydroxy-phenanthrene 1-Hydroxy-2-naphtho... |
|
map00470
|

|
D-Amino acid metabolism |
|
...)-1-carboxy-2-(1H-imidazol-4-yl)ethyl]amino}butanoate) C22024 (Staphylopine) C00049 (L-Aspartate) C0... |
....9 1-Pyrroline-2-carboxylate 5-Amino-2-oxo-pentanoate 2-Amino-4-oxo-pentanoate 2,4-Diamino-pentanoat... |
|
map00310
|
|
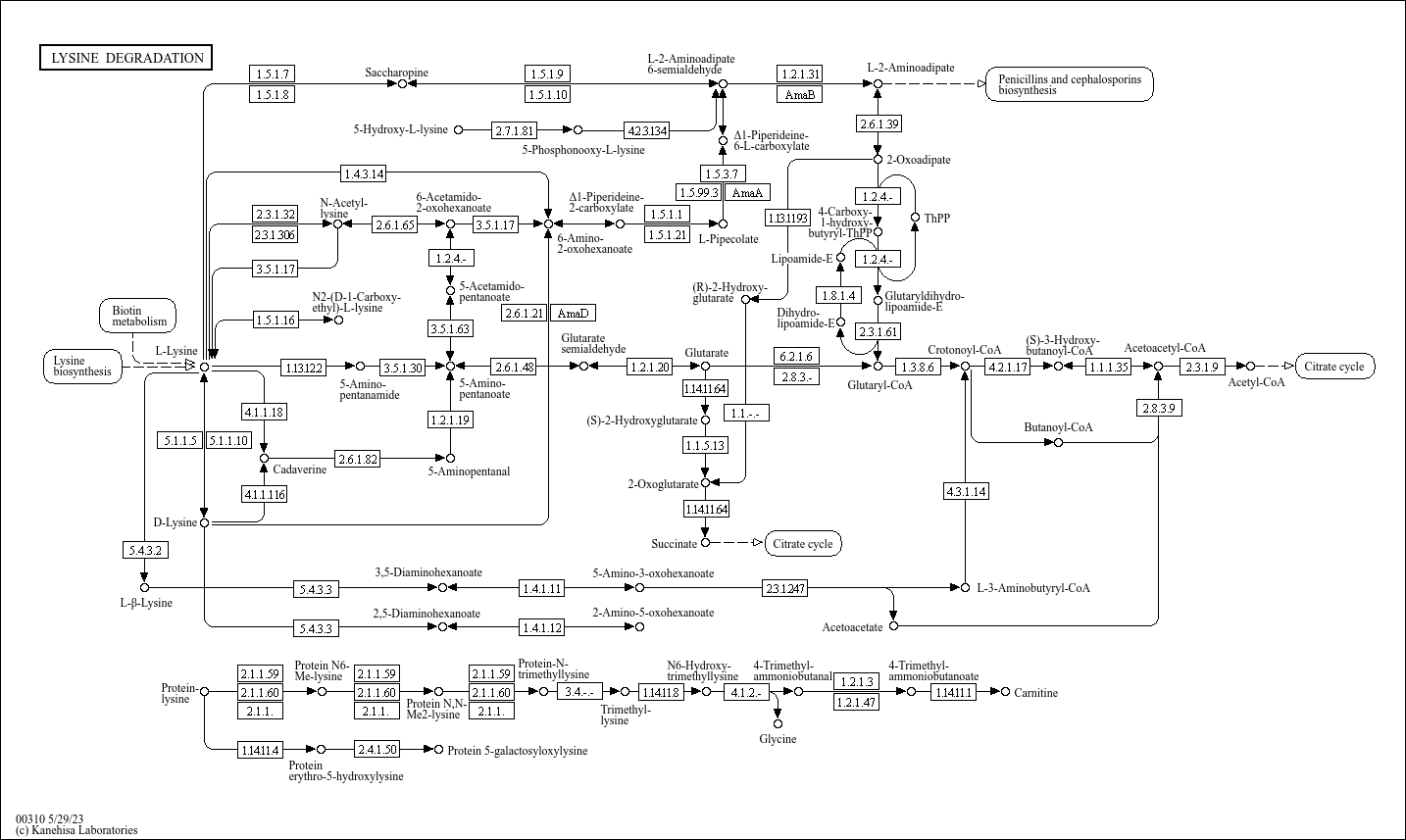
Lysine degradation |
|
C05825 (2-Amino-5-oxohexanoate) C05161 ((2R,5S)-2,5-Diaminohexanoate) C00877 (Crotonoyl-CoA) C01144 ... |
2.6.1.21 2-Amino-5-oxohexanoate 2,5-Diaminohexanoate 1.4.1.12 5.4.3.3 1.5.1.1 1.5.1.8 1.5.1.10 Penic... |