| Entry |
|
|---|
| Name |
Nimodipine (USP/INN);
Nimotop (TN);
Nymalize (TN) |
|---|
| Product |
|
|---|
| Generic |
|
|---|
| Formula |
C21H26N2O7
|
|---|
| Exact mass |
418.1740
|
|---|
| Mol weight |
418.44
|
|---|
| Structure |
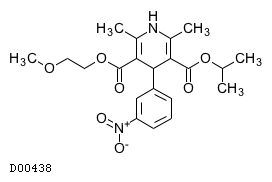
|
|---|
| Simcomp |
|
|---|
| Class |
Cardiovascular agent
DG01575 Calcium channel blocker
DG01496 Calcium channel L type blocker
DG03231 Antihypertensive
DG01928 Dihydropyridine calcium channel blocker
Metabolizing enzyme substrate
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
|
|---|
| Remark |
|
|---|
| Efficacy |
Antihypertensive, Vasodilator, Calcium channel blocker |
|---|
| Comment |
Dihydropyridine derivative
|
|---|
| Target |
|
|---|
| Pathway |
| hsa04261 | Adrenergic signaling in cardiomyocytes |
| hsa04270 | Vascular smooth muscle contraction |
|
|---|
| Metabolism |
Enzyme: CYP3A4 [HSA: 1576]
|
|---|
| Interaction |
|
|---|
| Structure map |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
C CARDIOVASCULAR SYSTEM
C08 CALCIUM CHANNEL BLOCKERS
C08C SELECTIVE CALCIUM CHANNEL BLOCKERS WITH MAINLY VASCULAR EFFECTS
C08CA Dihydropyridine derivatives
C08CA06 Nimodipine
D00438 Nimodipine (USP/INN) <US>
USP drug classification [BR:br08302]
Cardiovascular Agents
Calcium Channel Blocking Agents, Dihydropyridines
Nimodipine
D00438 Nimodipine (USP/INN)
Drug groups [BR:br08330]
Cardiovascular agent
DG01575 Calcium channel blocker
DG01496 Calcium channel L type blocker
D00438 Nimodipine
DG03231 Antihypertensive
DG01928 Dihydropyridine calcium channel blocker
D00438 Nimodipine
Metabolizing enzyme substrate
DG01633 CYP3A/CYP3A4 substrate
DG02913 CYP3A4 substrate
D00438 Nimodipine
Drug classes [BR:br08332]
Cardiovascular agent
DG01928 Dihydropyridine calcium channel blocker
D00438 Nimodipine
DG03231 Antihypertensive
D00438 Nimodipine
Target-based classification of drugs [BR:br08310]
Ion channels
Voltage-gated ion channels
Calcium channels
CACNA1-L
D00438 Nimodipine (USP/INN) <US>
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D00438
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 30
1 C1y C 26.2694 -15.6084
2 C8y C 26.2694 -17.0125
3 C2y C 25.0525 -14.9064
4 C2y C 27.4745 -14.9064
5 C8x C 25.0525 -17.7086
6 C8x C 27.4804 -17.7086
7 C2y C 25.0525 -13.5141
8 C7a C 23.8415 -15.6084
9 C2y C 27.4745 -13.5141
10 C7a C 28.6913 -15.6084
11 C8y C 25.0525 -19.1127
12 C8x C 27.4804 -19.1127
13 N1x N 26.2694 -12.8120
14 C1a C 23.8415 -12.8120
15 O7a O 22.6305 -14.9064
16 O6a O 23.8415 -17.0125
17 C1a C 28.6913 -12.8120
18 O7a O 29.9022 -14.9064
19 O6a O 28.6913 -17.0066
20 C8x C 26.2694 -19.8087
21 N2b N 23.8475 -19.8147 #+
22 C1b C 21.4195 -15.6084
23 C1c C 31.1132 -15.6084
24 O3a O 22.6305 -19.1127
25 O3a O 23.8475 -21.2070 #-
26 C1b C 20.2028 -14.9122
27 C1a C 32.3242 -14.9064
28 C1a C 31.0492 -17.0066
29 O2a O 18.9918 -15.6142
30 C1a C 17.7750 -14.9122
BOND 31
1 1 2 1
2 1 3 1
3 1 4 1
4 2 5 1
5 2 6 2
6 3 7 2
7 3 8 1
8 4 9 2
9 4 10 1
10 5 11 2
11 6 12 1
12 7 13 1
13 7 14 1
14 8 15 1
15 8 16 2
16 9 17 1
17 10 18 1
18 10 19 2
19 11 20 1
20 11 21 1
21 15 22 1
22 18 23 1
23 21 24 2
24 21 25 1
25 22 26 1
26 23 27 1
27 23 28 1
28 26 29 1
29 29 30 1
30 9 13 1
31 12 20 2
|
|---|