| Entry |
|
|---|
| Name |
Cytoskeleton in muscle cells - Drosophila melanogaster (fruit fly) |
|---|
| Class |
Cellular Processes; Cell motility
|
|---|
| Pathway map |
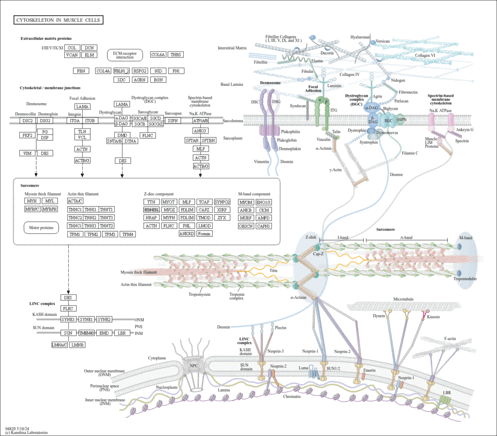
|
|---|
| Organism |
Drosophila melanogaster (fruit fly) [GN: dme]
|
|---|
| Gene |
| Dmel_CG14622 | DAAM; dishevelled associated activator of morphogenesis, isoform D [KO:K04512] |
| Dmel_CG30084 | Zasp52; Z band alternatively spliced PDZ-motif protein 52, isoform F [KO:K19867] |
|
|---|
| Reference |
|
|---|
| Authors |
Clark KA, McElhinny AS, Beckerle MC, Gregorio CC |
|---|
| Title |
Striated muscle cytoarchitecture: an intricate web of form and function. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mukund K, Subramaniam S |
|---|
| Title |
Skeletal muscle: A review of molecular structure and function, in health and disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Stroud MJ, Banerjee I, Veevers J, Chen J |
|---|
| Title |
Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC |
|---|
| Title |
Overview of the Muscle Cytoskeleton. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Csapo R, Gumpenberger M, Wessner B |
|---|
| Title |
Skeletal Muscle Extracellular Matrix - What Do We Know About Its Composition, Regulation, and Physiological Roles? A Narrative Review. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pinch M, Guth R, Samanta MP, Chaidez A, Unguez GA |
|---|
| Title |
The myogenic electric organ of Sternopygus macrurus: a non-contractile tissue with a skeletal muscle transcriptome. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sequeira V, Nijenkamp LL, Regan JA, van der Velden J |
|---|
| Title |
The physiological role of cardiac cytoskeleton and its alterations in heart failure. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang B, Powers JD, McCulloch AD, Chi NC |
|---|
| Title |
Nuclear mechanosignaling in striated muscle diseases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Al-Amoudi A, Castano-Diez D, Devos DP, Russell RB, Johnson GT, Frangakis AS |
|---|
| Title |
The three-dimensional molecular structure of the desmosomal plaque. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sikora M, Ermel UH, Seybold A, Kunz M, Calloni G, Reitz J, Vabulas RM, Hummer G, Frangakis AS |
|---|
| Title |
Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu XP, Kim E, Swift M, Smith JW, Volkmann N, Hanein D |
|---|
| Title |
Three-Dimensional Structures of Full-Length, Membrane-Embedded Human alpha(IIb)beta(3) Integrin Complexes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z |
|---|
| Title |
Structural insights into SUN-KASH complexes across the nuclear envelope. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sosa BA, Kutay U, Schwartz TU |
|---|
| Title |
Structural insights into LINC complexes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cruz VE, Esra Demircioglu F, Schwartz TU |
|---|
| Title |
Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Schuler SC, Liu Y, Dumontier S, Grandbois M, Le Moal E, Cornelison D, Bentzinger CF |
|---|
| Title |
Extracellular matrix: Brick and mortar in the skeletal muscle stem cell niche. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Brull A, Morales Rodriguez B, Bonne G, Muchir A, Bertrand AT |
|---|
| Title |
The Pathogenesis and Therapies of Striated Muscle Laminopathies. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Storey EC, Fuller HR |
|---|
| Title |
Genotype-Phenotype Correlations in Human Diseases Caused by Mutations of LINC Complex-Associated Genes: A Systematic Review and Meta-Summary. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|