| Entry |
|
|---|
| Name |
Oxidative phosphorylation - Haloplanus rubicundus CBA1112 |
|---|
| Class |
Metabolism; Energy metabolism
|
|---|
| Pathway map |
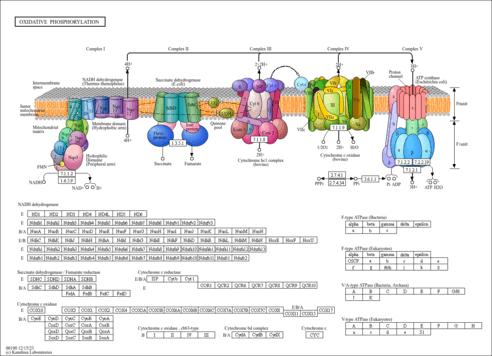
|
|---|
| Module |
|
|---|
| Organism |
Haloplanus rubicundus CBA1112 [GN: haq]
|
|---|
| Gene |
|
|---|
| Compound |
|
|---|
| Reference |
|
|---|
| Authors |
Sazanov LA, Hinchliffe P. |
|---|
| Title |
Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hinchliffe P, Carroll J, Sazanov LA. |
|---|
| Title |
Identification of a novel subunit of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|