| Entry |
|
|---|
| Name |
Oxidative phosphorylation - Mycobacterium tuberculosis variant bovis BCG Pasteur 1173P2 |
|---|
| Class |
Metabolism; Energy metabolism
|
|---|
| Pathway map |
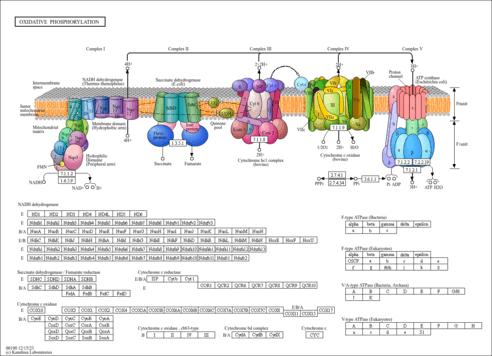
|
|---|
| Module |
|
|---|
| Organism |
Mycobacterium tuberculosis variant bovis BCG Pasteur 1173P2 [GN: mbb]
|
|---|
| Gene |
| BCG_1364 | atpB; Probable ATP synthase a chain atpB (protein 6) [KO:K02108] |
| BCG_1605 | frdBC; Probable fumarate reductase [iron-sulfur subunit] frdB and [membrane anchor subunit] frdC [KO:K00245 K00246] [EC:1.3.5.1] |
| BCG_1606 | frdD; Probable fumarate reductase [membrane anchor subunit] frdD [KO:K00247] |
| BCG_1660c | cydB; Probable integral membrane cytochrome D ubiquinol oxidase (subunit II) cydB [KO:K00426] [EC:7.1.1.7] |
| BCG_1661c | cydA; Probable integral membrane cytochrome D ubiquinol oxidase (subunit I) cydA [KO:K00425] [EC:7.1.1.7] |
| BCG_2210 | qcrC; Probable Ubiquinol-cytochrome C reductase QcrC (cytochrome C subunit) [KO:K03889] [EC:7.1.1.8] |
| BCG_2212 | qcrB; Probable Ubiquinol-cytochrome C reductase QcrB (cytochrome B subunit) [KO:K03891] [EC:7.1.1.8] |
| BCG_3382 | sdhC; Probable succinate dehydrogenase (cytochrome B-556 subunit) SDHC [KO:K00241] |
| BCG_3383 | sdhD; Probable succinate dehydrogenase (hydrophobic membrane anchor subunit) sdhD [KO:K00242] |
| BCG_3385 | sdhB; Probable succinate dehydrogenase (iron-sulphur protein subunit) sdhB [KO:K00240] [EC:1.3.5.1] |
|
|---|
| Compound |
|
|---|
| Reference |
|
|---|
| Authors |
Sazanov LA, Hinchliffe P. |
|---|
| Title |
Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hinchliffe P, Carroll J, Sazanov LA. |
|---|
| Title |
Identification of a novel subunit of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|