 | | PATHWAY: nau00770 | |
| Entry |
|
|---|
| Name |
Pantothenate and CoA biosynthesis - Nicotiana attenuata (coyote tobacco) |
|---|
| Class |
Metabolism; Metabolism of cofactors and vitamins
|
|---|
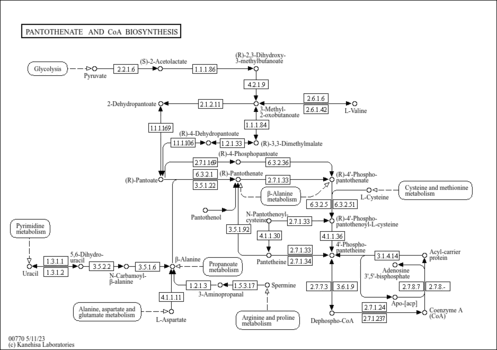
| Pathway map |
| nau00770 | Pantothenate and CoA biosynthesis |
|
|---|
| Module |
| nau_M00019 | Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine [PATH:nau00770] |
| nau_M00046 | Pyrimidine degradation, uracil => beta-alanine, thymine => 3-aminoisobutanoate [PATH:nau00770] |
|
|---|
| Other DBs |
|
|---|
| Organism |
Nicotiana attenuata (coyote tobacco) [GN: nau]
|
|---|
| Gene |
| 109229752 | uncharacterized protein LOC109229752 isoform X1 [KO:K06133] [EC:2.7.8.-] |
| 109234158 | uncharacterized protein LOC109234158 isoform X1 [KO:K06133] [EC:2.7.8.-] |
| 109212138 | L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [KO:K06133] [EC:2.7.8.-] |
|
|---|
| Compound |
| C00054 | Adenosine 3',5'-bisphosphate |
| C00141 | 3-Methyl-2-oxobutanoic acid |
| C01134 | Pantetheine 4'-phosphate |
| C03492 | D-4'-Phosphopantothenate |
| C03688 | Apo-[acyl-carrier protein] |
| C04079 | N-((R)-Pantothenoyl)-L-cysteine |
| C04272 | (R)-2,3-Dihydroxy-3-methylbutanoate |
| C04352 | (R)-4'-Phosphopantothenoyl-L-cysteine |
|
|---|
Related
pathway |
| nau00250 | Alanine, aspartate and glutamate metabolism |
| nau00270 | Cysteine and methionine metabolism |
| nau00330 | Arginine and proline metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|