 | | PATHWAY: fal00190 | |
| Entry |
|
|---|
| Name |
Oxidative phosphorylation - Frankia alni |
|---|
| Class |
Metabolism; Energy metabolism
|
|---|
| Pathway map |
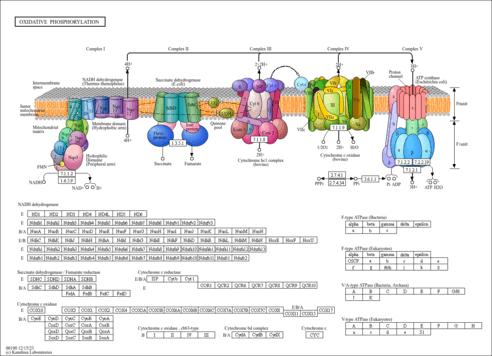
|
|---|
| Module |
|
|---|
| Organism |
|
|---|
| Gene |
| FRAAL1032 | nuoA; NADH-quinone oxidoreductase chain A (NADH dehydrogenase I, chain A) (NDH-1, chain A) [KO:K00330] [EC:7.1.1.2] |
| FRAAL1034 | nuoC; NADH-quinone oxidoreductase chain C (NADH dehydrogenase I, chain C) (NDH-1, chain C) [KO:K00332] [EC:7.1.1.2] |
| FRAAL1035 | nuoD; NADH-quinone oxidoreductase chain D (NADH dehydrogenase I, chain D) (NDH-1, chain D) [KO:K00333] [EC:7.1.1.2] |
| FRAAL1036 | nuoE; NADH-quinone oxidoreductase chain E (NADH dehydrogenase I, chain E) (NDH-1, chain E) [KO:K00334] [EC:7.1.1.2] |
| FRAAL1038 | nuoG; NADH-quinone oxidoreductase chain G (NADH dehydrogenase I, chain G) (NDH-1, chain G) [KO:K00336] [EC:7.1.1.2] |
| FRAAL1039 | nuoH; NADH-quinone oxidoreductase chain H (NADH dehydrogenase I, chain H) (NDH-1, chain H) [KO:K00337] [EC:7.1.1.2] |
| FRAAL1044 | nuoM; NADH-quinone oxidoreductase chain M (NADH dehydrogenase I, chain M) (NDH-1, chain M) [KO:K00342] [EC:7.1.1.2] |
| FRAAL1189 | sdhD; Succinate dehydrogenase hydrophobic membrane anchor protein [KO:K00242] |
| FRAAL5110 | qcrA; Ubiquinol-cytochrome c reductase iron-sulfur subunit (Rieske iron-sulfur protein) [KO:K03890] [EC:7.1.1.8] |
| FRAAL5932 | atpG; membrane-bound ATP synthase, F1 sector, gamma-subunit [KO:K02115] |
| FRAAL5930 | atpC; ATP synthase epsilon chain (ATP synthase F1 sector epsilon subunit) (partial) [KO:K02114] |
| FRAAL5826 | ppk; Polyphosphate kinase (Polyphosphoric acid kinase) (ATP-polyphosphate phosphotransferase) [KO:K00937] [EC:2.7.4.1] |
| FRAAL4412 | conserved hypothetical protein; putative P-loop containing nucleotide triphosphate hydrolase domain [KO:K22468] [EC:2.7.4.34] |
|
|---|
| Compound |
|
|---|
| Reference |
|
|---|
| Authors |
Sazanov LA, Hinchliffe P. |
|---|
| Title |
Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hinchliffe P, Carroll J, Sazanov LA. |
|---|
| Title |
Identification of a novel subunit of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|