| Entry |
|
|---|
| Name |
ATP-dependent chromatin remodeling - Nilaparvata lugens (brown planthopper) |
|---|
| Class |
Genetic Information Processing; Chromosome
|
|---|
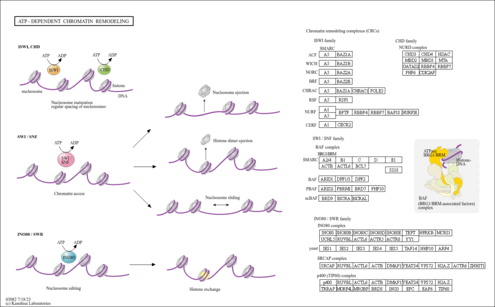
| Pathway map |
| nlu03082 | ATP-dependent chromatin remodeling |
|
|---|
| Organism |
Nilaparvata lugens (brown planthopper) [GN: nlu]
|
|---|
| Gene |
| 111048405 | chromatin-remodeling complex ATPase chain Iswi isoform X1 [KO:K11654] [EC:5.6.2.-] |
| 111049001 | bromodomain adjacent to zinc finger domain protein 1A isoform X1 [KO:K11655] |
| 111051023 | bromodomain adjacent to zinc finger domain protein 2B [KO:K26167] |
| 120348905 | chromatin accessibility complex protein 1-like isoform X1 [KO:K11656] |
| 111055473 | nucleosome-remodeling factor subunit NURF301 isoform X1 [KO:K11728] |
| 111053065 | chromodomain-helicase-DNA-binding protein Mi-2 homolog isoform X1 [KO:K11643] [EC:5.6.2.-] |
| 120349986 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1-like [KO:K11648] |
| 111046179 | LOW QUALITY PROTEIN: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 [KO:K11648] |
| 111061471 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 isoform X1 [KO:K11651] |
| 111051701 | LOW QUALITY PROTEIN: uncharacterized protein LOC111051701 [KO:K11671] |
| 111056925 | vacuolar protein sorting-associated protein 72 homolog isoform X1 [KO:K11664] |
| 111053487 | LOW QUALITY PROTEIN: transformation/transcription domain-associated protein [KO:K08874] |
|
|---|
| Reference |
|
|---|
| Authors |
Hasan N, Ahuja N |
|---|
| Title |
The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y |
|---|
| Title |
Structure of nucleosome-bound human BAF complex. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wanior M, Kramer A, Knapp S, Joerger AC |
|---|
| Title |
Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Burgio G, Onorati MC, Corona DF |
|---|
| Title |
Chromatin remodeling regulation by small molecules and metabolites. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|