| Entry |
|
|---|
| Name |
Motor proteins - Trichogramma pretiosum |
|---|
| Class |
Cellular Processes; Cell motility
|
|---|
| Pathway map |
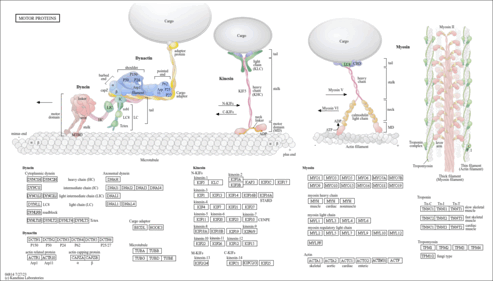
|
|---|
| Organism |
Trichogramma pretiosum [GN: tpre]
|
|---|
| Gene |
| 106656261 | cytoplasmic dynein 1 intermediate chain-like isoform X3 [KO:K10415] |
| 106649980 | cytoplasmic dynein 1 light intermediate chain 1 isoform X1 [KO:K10416] |
| 106650272 | transcription initiation factor TFIID subunit 12b-like [KO:K17914] |
|
|---|
| Reference |
|
|---|
| Authors |
Hirokawa N, Niwa S, Tanaka Y |
|---|
| Title |
Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Schmidt H, Zalyte R, Urnavicius L, Carter AP |
|---|
| Title |
Structure of human cytoplasmic dynein-2 primed for its power stroke. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G |
|---|
| Title |
The 2.8 A crystal structure of the dynein motor domain. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE |
|---|
| Title |
Structural basis for microtubule binding and release by dynein. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Gibbs KL, Greensmith L, Schiavo G |
|---|
| Title |
Regulation of Axonal Transport by Protein Kinases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, Bird AW, Carter AP |
|---|
| Title |
Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP |
|---|
| Title |
The structure of the dynactin complex and its interaction with dynein. |
|---|
| Journal |
|
|---|
| Reference |
PMID: 29420470 (PDB:5OWO 6F1T 6F1U 6F1V 6F1Y 6F1Z 6F38 6F3A) |
|---|
| Authors |
Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, Yildiz A, Carter AP |
|---|
| Title |
Cryo-EM shows how dynactin recruits two dyneins for faster movement. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Reck-Peterson SL, Redwine WB, Vale RD, Carter AP |
|---|
| Title |
The cytoplasmic dynein transport machinery and its many cargoes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H |
|---|
| Title |
Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C |
|---|
| Title |
Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E |
|---|
| Title |
The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM |
|---|
| Title |
Three-dimensional structure of myosin subfragment-1: a molecular motor. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Parker D, Bryant Z, Delp SL |
|---|
| Title |
Coarse-Grained Structural Modeling of Molecular Motors Using Multibody Dynamics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Dominguez R, Holmes KC |
|---|
| Title |
Actin structure and function. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yamada Y, Namba K, Fujii T |
|---|
| Title |
Cardiac muscle thin filament structures reveal calcium regulatory mechanism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kruppa AJ, Buss F |
|---|
| Title |
Motor proteins at the mitochondria-cytoskeleton interface. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|