| Entry |
|
|---|
| Name |
Efferocytosis - Homo sapiens (human) |
|---|
| Class |
Cellular Processes; Transport and catabolism
|
|---|
| Pathway map |
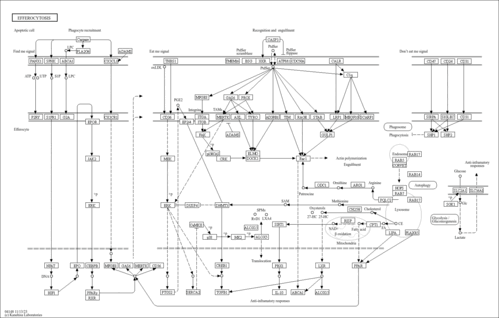
|
|---|
| Network |
|
|---|
| Element |
| N01747 | Find-me signal (nucleotide) |
| N01749 | Find-me signal (CX3CL1) |
| N01751 | Macrophage EPO signaling |
| N01753 | Exposure of phosphatidylserine to the outer leaflet |
| N01761 | Activation of CRK-DOCK-Rac1 pathway |
| N01762 | MERTK-mediated recognition and engulfment |
| N01763 | MEGF10-mediated recognition and engulfment |
| N01764 | Calreticulin-LRP1 mediated recognition and engulfment |
| N01773 | PTGS2-PGE2-TGFB1 pathway |
| N01774 | ERK-DUSP4 negative feedback pathway |
| N01775 | Inactivation of CaMKII by inducing SERCA2 |
| N01776 | CaMK2-p38-MK2-ALOX5 pathway |
| N01777 | Efferocytosis-induced NAD production |
| N01778 | Production of IL10 via the Sirtuin1 signaling cascade |
| N01779 | Continual efferocytosis enhanced by the AC-derived arginine and ornithine |
| N01780 | Hydrolyzing AC-derived cholesterol esters in the lysosome |
| N01781 | Activation of LXRs by oxysterols |
| N01784 | Glucose uptake and lactate release induced by efferocytosis |
| N01785 | Don't eat me signal (CD47) |
| N01786 | Don't eat me signal (CD24) |
|
|---|
| Drug |
| D10931 | Etrasimod arginine (USAN) |
| D12772 | Mocravimod hydrochloride (USAN) |
| D12798 | Verzistobart (USAN/INN) |
| D12799 | Vociprotafib (USAN/INN) |
| D12826 | Maplirpacept (USAN/INN) |
| D12892 | Migoprotafib (USAN/INN) |
|
|---|
| Organism |
Homo sapiens (human) [GN: hsa]
|
|---|
| Gene |
| 5031 | P2RY6; pyrimidinergic receptor P2Y6 [KO:K04272] |
| 1901 | S1PR1; sphingosine-1-phosphate receptor 1 [KO:K04288] |
| 19 | ABCA1; ATP binding cassette subfamily A member 1 [KO:K05641] |
| 29933 | GPR132; G protein-coupled receptor 132 [KO:K08426] |
| 6376 | CX3CL1; C-X3-C motif chemokine ligand 1 [KO:K05508] |
| 1524 | CX3CR1; C-X3-C motif chemokine receptor 1 [KO:K04192] |
| 4773 | NFATC2; nuclear factor of activated T cells 2 [KO:K17332] |
| 4772 | NFATC1; nuclear factor of activated T cells 1 [KO:K04446] |
| 4775 | NFATC3; nuclear factor of activated T cells 3 [KO:K17333] |
| 4776 | NFATC4; nuclear factor of activated T cells 4 [KO:K17334] |
| 3091 | HIF1A; hypoxia inducible factor 1 subunit alpha [KO:K08268] |
| 405 | ARNT; aryl hydrocarbon receptor nuclear translocator [KO:K09097] |
| 1051 | CEBPB; CCAAT enhancer binding protein beta [KO:K10048] |
| 5468 | PPARG; peroxisome proliferator activated receptor gamma [KO:K08530] |
| 4240 | MFGE8; milk fat globule EGF and factor V/VIII domain containing [KO:K17253] |
| 948 | CD36; CD36 molecule (CD36 blood group) [KO:K06259] |
| 5732 | PTGER2; prostaglandin E receptor 2 [KO:K04259] |
| 5734 | PTGER4; prostaglandin E receptor 4 [KO:K04261] |
| 1385 | CREB1; cAMP responsive element binding protein 1 [KO:K05870] |
| 7040 | TGFB1; transforming growth factor beta 1 [KO:K13375] |
| 487 | ATP2A1; ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 [KO:K05853] [EC:7.2.2.10] |
| 489 | ATP2A3; ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 [KO:K05853] [EC:7.2.2.10] |
| 488 | ATP2A2; ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 [KO:K05853] [EC:7.2.2.10] |
| 815 | CAMK2A; calcium/calmodulin dependent protein kinase II alpha [KO:K04515] [EC:2.7.11.17] |
| 817 | CAMK2D; calcium/calmodulin dependent protein kinase II delta [KO:K04515] [EC:2.7.11.17] |
| 816 | CAMK2B; calcium/calmodulin dependent protein kinase II beta [KO:K04515] [EC:2.7.11.17] |
| 818 | CAMK2G; calcium/calmodulin dependent protein kinase II gamma [KO:K04515] [EC:2.7.11.17] |
| 682 | BSG; basigin (Ok blood group) [KO:K06535] |
| 575 | ADGRB1; adhesion G protein-coupled receptor B1 [KO:K04596] |
| 91937 | TIMD4; T cell immunoglobulin and mucin domain containing 4 [KO:K26936] |
| 26762 | HAVCR1; hepatitis A virus cellular receptor 1 [KO:K20413] |
| 84868 | HAVCR2; hepatitis A virus cellular receptor 2 [KO:K20414] |
| 177 | AGER; advanced glycosylation end-product specific receptor [KO:K19722] |
| 9564 | BCAR1; BCAR1 scaffold protein, Cas family member [KO:K05726] |
| 1398 | CRK; CRK proto-oncogene, adaptor protein [KO:K04438] |
| 1399 | CRKL; CRK like proto-oncogene, adaptor protein [KO:K04438] |
| 9844 | ELMO1; engulfment and cell motility 1 [KO:K12366] |
| 1793 | DOCK1; dedicator of cytokinesis 1 [KO:K13708] |
| 8754 | ADAM9; ADAM metallopeptidase domain 9 [KO:K06834] [EC:3.4.24.-] |
| 4035 | LRP1; LDL receptor related protein 1 [KO:K04550] |
| 8578 | SCARF1; scavenger receptor class F member 1 [KO:K24318] |
| 51454 | GULP1; GULP PTB domain containing engulfment adaptor 1 [KO:K23285] |
| 64284 | RAB17; RAB17, member RAS oncogene family [KO:K07909] |
| 5868 | RAB5A; RAB5A, member RAS oncogene family [KO:K07887] |
| 5869 | RAB5B; RAB5B, member RAS oncogene family [KO:K07888] |
| 5878 | RAB5C; RAB5C, member RAS oncogene family [KO:K07889] |
| 9392 | TGFBRAP1; transforming growth factor beta receptor associated protein 1 [KO:K20177] |
| 55823 | VPS11; VPS11 core subunit of CORVET and HOPS complexes [KO:K20179] |
| 64601 | VPS16; VPS16 core subunit of CORVET and HOPS complexes [KO:K20180] |
| 57617 | VPS18; VPS18 core subunit of CORVET and HOPS complexes [KO:K20181] |
| 65082 | VPS33A; VPS33A core subunit of CORVET and HOPS complexes [KO:K20182] |
| 51552 | RAB14; RAB14, member RAS oncogene family [KO:K07881] |
| 7879 | RAB7A; RAB7A, member RAS oncogene family [KO:K07897] |
| 54896 | SLC66A1; solute carrier family 66 member 1 [KO:K23678] |
| 10062 | NR1H3; nuclear receptor subfamily 1 group H member 3 [KO:K08536] |
| 7376 | NR1H2; nuclear receptor subfamily 1 group H member 2 [KO:K08535] |
| 7386 | UQCRFS1; ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 [KO:K00411] [EC:7.1.1.8] |
| 5467 | PPARD; peroxisome proliferator activated receptor delta [KO:K04504] |
| 6513 | SLC2A1; solute carrier family 2 member 1 [KO:K07299] |
| 6566 | SLC16A1; solute carrier family 16 member 1 [KO:K08179] |
| 5175 | PECAM1; platelet and endothelial cell adhesion molecule 1 [KO:K06471] |
| 89790 | SIGLEC10; sialic acid binding Ig like lectin 10 [KO:K06749] |
| 55423 | SIRPG; signal regulatory protein gamma [KO:K06551] |
| 10326 | SIRPB1; signal regulatory protein beta 1 [KO:K06551] |
|
|---|
| Compound |
| C00019 | S-Adenosyl-L-methionine |
| C04230 | 1-Acyl-sn-glycero-3-phosphocholine |
| C06124 | Sphingosine 1-phosphate |
| C15610 | Cholest-5-ene-3beta,26-diol |
|
|---|
| Reference |
|
|---|
| Authors |
Doran AC, Yurdagul A Jr, Tabas I |
|---|
| Title |
Efferocytosis in health and disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang J, Ding W, Zhao M, Liu J, Xu Y, Wan J, Wang M |
|---|
| Title |
Mechanisms of efferocytosis in determining inflammation resolution: Therapeutic potential and the association with cardiovascular disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kawano M, Nagata S |
|---|
| Title |
Efferocytosis and autoimmune disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mehrotra P, Ravichandran KS |
|---|
| Title |
Drugging the efferocytosis process: concepts and opportunities. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Park SY, Kim IS |
|---|
| Title |
Engulfment signals and the phagocytic machinery for apoptotic cell clearance. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kojima Y, Weissman IL, Leeper NJ |
|---|
| Title |
The Role of Efferocytosis in Atherosclerosis. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|