| Entry |
|
|---|
| Name |
Enzyme N6-(dihydrolipoyl)lysine;
Dihydrolipoamide-E;
[E2 protein]-N6-[(R)-dihydrolipoyl]-L-lysine;
[Lipoyl-carrier protein E2]-N6-[(R)-dihydrolipoyl]-L-lysine
|
|---|
| Formula |
C8H16NOS2R
|
|---|
| Structure |
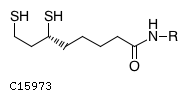
|
|---|
| Comment |
Generic compound in reaction hierarchy
The reduced lipoyllysine residue in EC 2.3.1.12, dihydrolipoyllysine-residue acetyltransferase ( EC 2.3.1.61, dihydrolipoyllysine-residue succinyltransferase or EC 2.3.1.168, dihydrolipoyllysine-residue (2-methylpropanoyl)transferase ).
|
|---|
| Reaction |
|
|---|
| Pathway |
| map00280 | Valine, leucine and isoleucine degradation |
|
|---|
| Module |
| M00009 | Citrate cycle (TCA cycle, Krebs cycle) |
| M00011 | Citrate cycle, second carbon oxidation, 2-oxoglutarate => oxaloacetate |
| M00032 | Lysine degradation, lysine => saccharopine => acetoacetyl-CoA |
| M00036 | Leucine degradation, leucine => acetoacetate + acetyl-CoA |
| M00307 | Pyruvate oxidation, pyruvate => acetyl-CoA |
| M00881 | Lipoic acid biosynthesis, plants and bacteria, octanoyl-ACP => dihydrolipoyl-E2/H |
| M00883 | Lipoic acid biosynthesis, animals and bacteria, octanoyl-ACP => dihydrolipoyl-H => dihydrolipoyl-E2 |
| M00884 | Lipoic acid biosynthesis, octanoyl-CoA => dihydrolipoyl-E2 |
|
|---|
| Network |
nt06024 Valine, leucine and isoleucine degradation nt06031 Citrate cycle and pyruvate metabolism nt06032 Lipoic acid metabolism nt06036 Lysine degradation |
|---|
| Enzyme |
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 13
1 C1c C 15.4683 -15.8087
2 C1b C 16.6705 -16.5090
3 C1b C 14.2660 -16.5090
4 S1a S 15.4683 -14.4255
5 C1b C 17.8728 -15.8087
6 C1b C 13.0639 -15.8087
7 C1b C 19.0808 -16.5090
8 S1a S 13.0639 -14.4255
9 C1b C 20.2771 -15.8087
10 C5a C 21.4794 -16.5090
11 O5a O 21.4794 -17.8921
12 N1b N 22.8334 -15.8379
13 R R 24.2335 -15.8379
BOND 12
1 1 2 1 #Down
2 1 3 1
3 1 4 1
4 2 5 1
5 3 6 1
6 5 7 1
7 6 8 1
8 7 9 1
9 9 10 1
10 10 11 2
11 10 12 1
12 12 13 1
|
|---|