| Entry |
|
|---|
| Name |
Fructose and mannose metabolism - Gemmata massiliana |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
| gms00051 | Fructose and mannose metabolism |
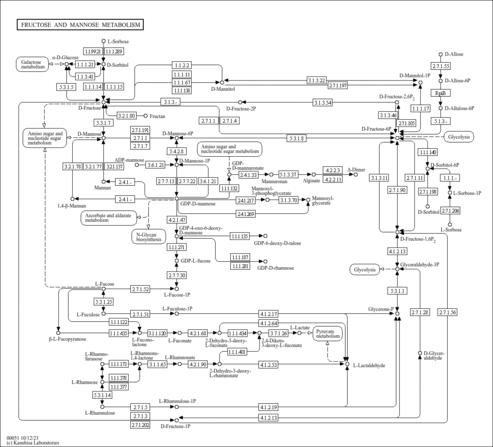
|
|---|
| Other DBs |
|
|---|
| Organism |
Gemmata massiliana [GN: gms]
|
|---|
| Gene |
| SOIL9_58960 | carbohydrate family : Kinase, PfkB family OS=Porphyromonas sp. oral taxon 278 str. W7784 GN=HMPREF1556_00125 PE=4 SV=1: PfkB [KO:K00847] [EC:2.7.1.4] |
| SOIL9_00430 | mannose-6-phosphate isomerase : Phosphomannose isomerase OS=Singulisphaera acidiphila (strain ATCC BAA-1392 / DSM 18658 / VKM B-2454 / MOB10) GN=Sinac_2015 PE=4 SV=1: PMI_typeI [KO:K01809] [EC:5.3.1.8] |
| SOIL9_17130 | phosphomannomutase : Phosphoglucomutase/phosphomannomutase family protein OS=Rhodopirellula europaea 6C GN=RE6C_01147 PE=3 SV=1: PGM_PMM_I: PGM_PMM_II: PGM_PMM_III: PGM_PMM_IV [KO:K01840] [EC:5.4.2.8] |
| SOIL9_40190 | mannose-1-phosphate guanylyltransferase : Mannose-1-phosphate guanylyltransferase OS=Blastopirellula marina DSM 3645 GN=DSM3645_16810 PE=4 SV=1: NTP_transferase: MannoseP_isomer [KO:K00971] [EC:2.7.7.13] |
| SOIL9_45910 | nucleoside-diphosphate-sugar pyrophosphorylase : Nucleotidyl transferase OS=Oribacterium sinus F0268 GN=HMPREF6123_1285 PE=4 SV=1: NTP_transferase [KO:K00966] [EC:2.7.7.13] |
| SOIL9_38280 | mannose-1-phosphate guanyltransferase : Uncharacterized protein OS=uncultured bacterium GN=ACD_20C00350G0017 PE=4 SV=1: NTP_transferase [KO:K00966] [EC:2.7.7.13] |
| SOIL9_61700 | gdp-mannose -dehydratase : GDP-mannose 4,6-dehydratase OS=Sphaerobacter thermophilus (strain DSM 20745 / S 6022) GN=gmd PE=3 SV=1: Epimerase [KO:K01711] [EC:4.2.1.47] |
| SOIL9_46320 | nad-dependent dehydratase : GDP-L-fucose synthase OS=Thermosynechococcus sp. NK55a GN=wcaG PE=3 SV=1: Epimerase [KO:K02377] [EC:1.1.1.271] |
| SOIL9_47670 | nad-dependent dehydratase : Marine sediment metagenome DNA, contig: S01H1_S31473 (Fragment) OS=marine sediment metagenome GN=S01H1_69638 PE=3 SV=1: Epimerase [KO:K02377] [EC:1.1.1.271] |
| SOIL9_53930 | nad-dependent dehydratase : GDP-L-fucose synthase OS=Brevibacillus panacihumi W25 GN=fcl PE=3 SV=1: Epimerase [KO:K02377] [EC:1.1.1.271] |
| SOIL9_33220 | gdp-l-fucose synthase : GDP-L-fucose synthase OS=Anoxybacillus flavithermus AK1 GN=fcl PE=3 SV=1: Epimerase [KO:K02377] [EC:1.1.1.271] |
| SOIL9_49820 | short-chain alcohol dehydrogenase : (S)-1-phenylethanol dehydrogenase Ped OS=Methyloglobulus morosus KoM1 GN=ped PE=3 SV=1: adh_short [KO:K21883] [EC:1.1.1.401] |
| SOIL9_18330 | 3-oxoacyl-acp reductase : Uncharacterized protein OS=Singulisphaera acidiphila (strain ATCC BAA-1392 / DSM 18658 / VKM B-2454 / MOB10) GN=Sinac_5872 PE=3 SV=1: adh_short [KO:K21883] [EC:1.1.1.401] |
| SOIL9_76860 | xylose isomerase : Xylose isomerase OS=Blastopirellula marina DSM 3645 GN=xylA PE=3 SV=1: AP_endonuc_2 [KO:K01805] [EC:5.3.1.5] |
| SOIL9_59720 | triosephosphate isomerase : Triosephosphate isomerase OS=Pelotomaculum thermopropionicum (strain DSM 13744 / JCM 10971 / SI) GN=TpiA PE=3 SV=1: TIM [KO:K01803] [EC:5.3.1.1] |
| SOIL9_64440 | glycosyl hydrolase family 32 : Beta-fructosidase, levanase/invertase OS=Singulisphaera acidiphila (strain ATCC BAA-1392 / DSM 18658 / VKM B-2454 / MOB10) GN=Sinac_4623 PE=4 SV=1: Glyco_hydro_32N: Glyco_hydro_32C [KO:K03332] [EC:3.2.1.80] |
| SOIL9_61690 | Marine sediment metagenome DNA, contig: S03H2_L04007 (Fragment) OS=marine sediment metagenome GN=S03H2_17566 PE=4 SV=1: Epimerase [KO:K15856] [EC:1.1.1.281] |
| SOIL9_62440 | ribose 5-phosphate isomerase : Ribose-5-phosphate isomerase OS=Planctomyces brasiliensis (strain ATCC 49424 / DSM 5305 / JCM 21570 / NBRC 103401 / IFAM 1448) GN=Plabr_1998 PE=4 SV=1: LacAB_rpiB [KO:K01808] [EC:5.3.1.6] |
|
|---|
| Compound |
| C00118 | D-Glyceraldehyde 3-phosphate |
| C00354 | D-Fructose 1,6-bisphosphate |
| C00665 | beta-D-Fructose 2,6-bisphosphate |
| C01131 | L-Rhamnulose 1-phosphate |
| C01222 | GDP-4-dehydro-6-deoxy-D-mannose |
| C03267 | beta-D-Fructose 2-phosphate |
| C03827 | 2-Dehydro-3-deoxy-L-fuconate |
| C03979 | 2-Dehydro-3-deoxy-L-rhamnonate |
| C05392 | Oligouronide with 4-deoxy-alpha-L-erythro-hex-4-enopyranuronosyl group |
| C11516 | 2-(alpha-D-Mannosyl)-3-phosphoglycerate |
| C11544 | 2-O-(alpha-D-Mannosyl)-D-glycerate |
| C20781 | 2,4-Diketo-3-deoxy-L-fuconate |
|
|---|
Related
pathway |
| gms00053 | Ascorbate and aldarate metabolism |
| gms00520 | Amino sugar and nucleotide sugar metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|