| Entry |
|
|---|
| Name |
ATP-dependent chromatin remodeling - Haliaeetus leucocephalus (bald eagle) |
|---|
| Class |
Genetic Information Processing; Chromosome
|
|---|
| Pathway map |
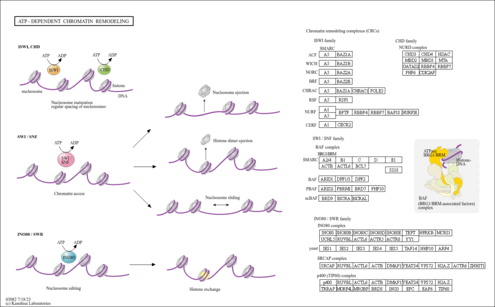
| hle03082 | ATP-dependent chromatin remodeling |
|
|---|
| Organism |
Haliaeetus leucocephalus (bald eagle) [GN: hle]
|
|---|
| Gene |
| 104831397 | SMARCA5; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 [KO:K11654] [EC:5.6.2.-] |
| 104841769 | BAZ1A; bromodomain adjacent to zinc finger domain protein 1A isoform X1 [KO:K11655] |
| 104830306 | BAZ2A; LOW QUALITY PROTEIN: bromodomain adjacent to zinc finger domain protein 2A [KO:K15224] |
| 104832870 | BAZ2B; bromodomain adjacent to zinc finger domain protein 2B isoform X1 [KO:K26167] |
| 104832682 | SMARCA1; probable global transcription activator SNF2L1 isoform X1 [KO:K11727] [EC:5.6.2.-] |
| 104838723 | BPTF; LOW QUALITY PROTEIN: nucleosome-remodeling factor subunit BPTF [KO:K11728] |
| 104834246 | CHD3; chromodomain-helicase-DNA-binding protein 3 [KO:K11642] [EC:5.6.2.-] |
| 104831755 | CHD4; chromodomain-helicase-DNA-binding protein 4 isoform X1 [KO:K11643] [EC:5.6.2.-] |
| 104837423 | MTA3; LOW QUALITY PROTEIN: metastasis-associated protein MTA3 [KO:K11660] |
| 104829120 | GATAD2A; transcriptional repressor p66-alpha isoform X1 [KO:K23194] |
| 104840915 | CDK2AP1; cyclin-dependent kinase 2-associated protein 1 [KO:K26454] |
| 104834103 | SMARCA4; transcription activator BRG1 isoform X1 [KO:K11647] [EC:5.6.2.-] |
| 104841248 | SMARCA2; probable global transcription activator SNF2L2 isoform X1 [KO:K11647] [EC:5.6.2.-] |
| 104836775 | SMARCB1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 isoform X1 [KO:K11648] |
| 104827968 | SMARCD2; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 [KO:K11650] |
| 104834314 | SMARCD3; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 [KO:K11650] |
| 104831134 | SMARCD1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [KO:K11650] |
| 104842914 | SMARCE1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 [KO:K11651] |
| 104841295 | AGFG2; arf-GAP domain and FG repeat-containing protein 2 [KO:K11652] |
| 104843168 | BCL7B; B-cell CLL/lymphoma 7 protein family member B isoform X1 [KO:K25605] |
| 104840887 | BCL7A; B-cell CLL/lymphoma 7 protein family member A isoform X1 [KO:K25605] |
| 104837190 | ARID1A; AT-rich interactive domain-containing protein 1A [KO:K11653] |
| 104840705 | ARID1B; AT-rich interactive domain-containing protein 1B isoform X1 [KO:K11653] |
| 104835854 | ARID2; AT-rich interactive domain-containing protein 2 [KO:K11765] |
| 104842585 | BRD7; LOW QUALITY PROTEIN: bromodomain-containing protein 7 [KO:K11723] |
| 104837622 | BRD9; LOW QUALITY PROTEIN: bromodomain-containing protein 9 [KO:K22184] |
| 104831501 | GLTSCR1; glioma tumor suppressor candidate region gene 1 protein isoform X1 [KO:K25612] |
| 104840455 | NFRKB; nuclear factor related to kappa-B-binding protein [KO:K11671] |
| 104841914 | YY1; transcriptional repressor protein YY1 isoform X1 [KO:K09201] |
| 104841073 | DMAP1; DNA methyltransferase 1-associated protein 1 isoform X1 [KO:K11324] |
| 104830572 | VPS72; vacuolar protein sorting-associated protein 72 homolog [KO:K11664] |
| 104840631 | EP400; E1A-binding protein p400 isoform X1 [KO:K11320] [EC:5.6.2.-] |
| 104833531 | TRRAP; transformation/transcription domain-associated protein isoform X1 [KO:K08874] |
| 104834634 | MORF4L1; mortality factor 4-like protein 1 isoform X1 [KO:K11339] |
|
|---|
| Reference |
|
|---|
| Authors |
Hasan N, Ahuja N |
|---|
| Title |
The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y |
|---|
| Title |
Structure of nucleosome-bound human BAF complex. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Wanior M, Kramer A, Knapp S, Joerger AC |
|---|
| Title |
Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Burgio G, Onorati MC, Corona DF |
|---|
| Title |
Chromatin remodeling regulation by small molecules and metabolites. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|