| Entry |
|
|---|
| Name |
Valine, leucine and isoleucine biosynthesis - Oceanisphaera avium |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
| ocm00290 | Valine, leucine and isoleucine biosynthesis |
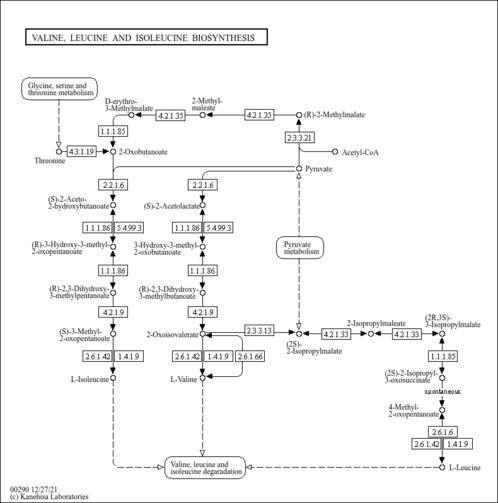
|
|---|
| Module |
| ocm_M00019 | Valine/isoleucine biosynthesis, pyruvate => valine / 2-oxobutanoate => isoleucine [PATH:ocm00290] |
| ocm_M00570 | Isoleucine biosynthesis, threonine => 2-oxobutanoate => isoleucine [PATH:ocm00290] |
|
|---|
| Other DBs |
|
|---|
| Organism |
Oceanisphaera avium [GN: ocm]
|
|---|
| Gene |
|
|---|
| Compound |
| C00141 | 3-Methyl-2-oxobutanoic acid |
| C00233 | 4-Methyl-2-oxopentanoate |
| C00671 | (S)-3-Methyl-2-oxopentanoic acid |
| C04181 | 3-Hydroxy-3-methyl-2-oxobutanoic acid |
| C04236 | (2S)-2-Isopropyl-3-oxosuccinate |
| C04272 | (R)-2,3-Dihydroxy-3-methylbutanoate |
| C04411 | (2R,3S)-3-Isopropylmalate |
| C06006 | (S)-2-Aceto-2-hydroxybutanoate |
| C06007 | (R)-2,3-Dihydroxy-3-methylpentanoate |
| C06032 | D-erythro-3-Methylmalate |
| C14463 | (R)-3-Hydroxy-3-methyl-2-oxopentanoate |
|
|---|
| Reference |
|
|---|
| Authors |
Xu H, Zhang Y, Guo X, Ren S, Staempfli AA, Chiao J, Jiang W, Zhao G. |
|---|
| Title |
Isoleucine biosynthesis in Leptospira interrogans serotype lai strain 56601 proceeds via a threonine-independent pathway. |
|---|
| Journal |
|
|---|
Related
pathway |
| ocm00260 | Glycine, serine and threonine metabolism |
| ocm00280 | Valine, leucine and isoleucine degradation |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|