| Entry |
|
|---|
| Name |
Alanine, aspartate and glutamate metabolism - Sedimentisphaera salicampi |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
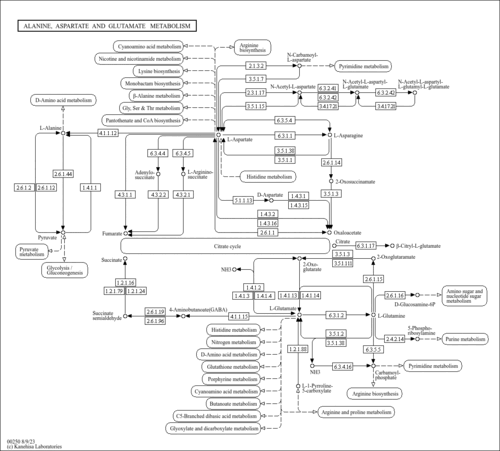
| Pathway map |
| pbp00250 | Alanine, aspartate and glutamate metabolism |
|
|---|
| Other DBs |
|
|---|
| Organism |
Sedimentisphaera salicampi [GN: pbp]
|
|---|
| Gene |
|
|---|
| Compound |
| C00352 | D-Glucosamine 6-phosphate |
| C00438 | N-Carbamoyl-L-aspartate |
| C03406 | N-(L-Arginino)succinate |
| C03794 | N6-(1,2-Dicarboxyethyl)-AMP |
| C03912 | (S)-1-Pyrroline-5-carboxylate |
| C12270 | N-Acetylaspartylglutamate |
| C20775 | beta-Citryl-L-glutamate |
| C20776 | N-Acetylaspartylglutamylglutamate |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Wu G |
|---|
| Title |
Intestinal mucosal amino acid catabolism. |
|---|
| Journal |
|
|---|
Related
pathway |
| pbp00260 | Glycine, serine and threonine metabolism |
| pbp00330 | Arginine and proline metabolism |
| pbp00520 | Amino sugar and nucleotide sugar metabolism |
| pbp00630 | Glyoxylate and dicarboxylate metabolism |
| pbp00660 | C5-Branched dibasic acid metabolism |
| pbp00760 | Nicotinate and nicotinamide metabolism |
| pbp00770 | Pantothenate and CoA biosynthesis |
|
|---|
| KO pathway |
|
|---|