| Entry |
|
|---|
| Name |
Sulfur metabolism |
|---|
| Description |
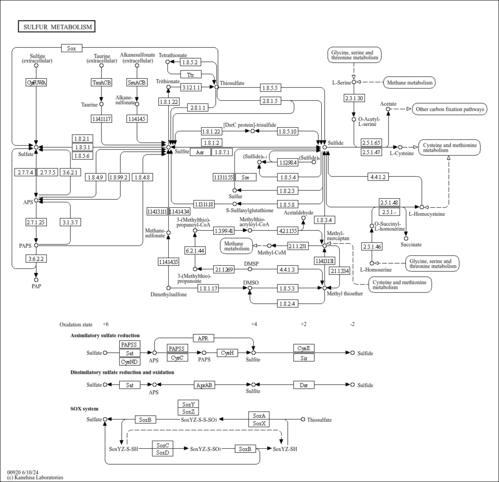
Sulfur is an essential element for life and the metabolism of organic sulfur compounds plays an important role in the global sulfur cycle. Sulfur occurs in various oxidation states ranging from +6 in sulfate to -2 in sulfide (H2S). Sulfate reduction can occur in both an energy consuming assimilatory pathway and an energy producing dissimilatory pathway. The assimilatory pathway, which is found in a wide range of organisms, produces reduced sulfur compounds for the biosynthesis of S-containing amino acids and does not lead to direct excretion of sulfide. In the dissimilatory pathway, which is restricted to obligatory anaerobic bacterial and archaeal lineages, sulfate (or sulfur) is the terminal electron acceptor of the respiratory chain producing large quantities of inorganic sulfide. Both pathways start from the activation of sulfate by reaction with ATP to form adenylyl sulfate (APS). In the assimilatory pathway [MD: M00176] APS is converted to 3'-phosphoadenylyl sulfate (PAPS) and then reduced to sulfite, and sulfite is further reduced to sulfide by the assimilatory sulfite reductase. In the dissimilatory pathway [MD: M00596] APS is directly reduced to sulfite, and sulfite is further reduced to sulfide by the dissimilatory sulfite reductase. The capacity for oxidation of sulfur is quite widespread among bacteria and archaea, comprising phototrophs and chemolithoautotrophs. The SOX (sulfur-oxidation) system [MD: M00595] is a well-known sulfur oxidation pathway and is found in both photosynthetic and non-photosynthetic sulfur-oxidizing bacteria. Green sulfur bacteria and purple sulfur bacteria carry out anoxygenic photosynthesis with reduced sulfur compounds such as sulfide and elemental sulfur, as well as thiosulfate (in some species with the SOX system), as the electron donor for photoautotrophic growth. In some chemolithoautotrophic sulfur oxidizers (such as Thiobacillus denitrificans), it has been suggested that dissimilatory sulfur reduction enzymes operate in the reverse direction, forming a sulfur oxidation pathway from sulfite to APS and then to sulfate.
|
|---|
| Class |
Metabolism; Energy metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| M00176 | Assimilatory sulfate reduction, sulfate => H2S [PATH:rn00920] |
| M00595 | Sulfur oxidation, SOX system, thiosulfate => sulfate [PATH:rn00920] |
| M00596 | Dissimilatory sulfate reduction, sulfate => H2S [PATH:rn00920] |
| M00984 | Sulfur oxidation, tetrathionate intermediate (S4I) pathway, thiosulfate => sulfur + sulfate + thiosulfate [PATH:rn00920] |
| M00987 | Assimilatory sulfate reduction, plants, sulfate => H2S [PATH:rn00920] |
| M00990 | Dimethylsulfoniopropionate (DMSP) degradation, demethylation pathway, DMSP => methanethiol [PATH:rn00920] |
| M00991 | Dimethylsulfoniopropionate (DMSP) degradation, cleavage pathway, DMSP => acetyl-CoA [PATH:rn00920] |
| M00992 | Dimethylsulfoniopropionate (DMSP) degradation, cleavage pathway, DMSP => acrylate => acetyl-CoA [PATH:rn00920] |
| M00993 | Dimethylsulfoniopropionate (DMSP) degradation, cleavage pathway, DMSP => propionyl-CoA [PATH:rn00920] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00029 | thiosulfate:ferricytochrome-c oxidoreductase |
| R00295 | trithionate,NAD+ oxidoreductase |
| R00507 | 3'-phosphoadenylyl-sulfate sulfohydrolase; 3'-phosphoadenylyl-sulfate nucleotidohydrolase |
| R00508 | 3'-phospho-5'-adenylyl sulfate 3'-phosphohydrolase |
| R00509 | ATP:adenylylsulfate 3'-phosphotransferase |
| R00528 | sulfite:ferricytochrome-c oxidoreductase |
| R00529 | ATP:sulfate adenylyltransferase |
| R00530 | ADP:sulfate adenylyltransferase |
| R00531 | adenylylsulfate sulfohydrolase |
| R00533 | sulfite:oxygen oxidoreductase |
| R00586 | acetyl-CoA:L-serine O-acetyltransferase |
| R00649 | S-adenosyl-L-methionine:L-methionine S-methyltransferase |
| R00858 | hydrogen-sulfide:NADP+ oxidoreductase |
| R00859 | hydrogen-sulfide:ferredoxin oxidoreductase |
| R00861 | [DsrC protein]-trisulfide,NAD+ oxidoreductase (sulfite-forming) |
| R00897 | O3-acetyl-L-serine:hydrogen-sulfide 2-amino-2-carboxyethyltransferase; O3-acetyl-L-serine acetate-lyase (adding hydrogen sulfide) |
| R01283 | L-homocysteine hydrogen-sulfide-lyase (deaminating; 2-oxobutanoate-forming) |
| R01288 | O4-succinyl-L-homoserine:hydrogen sulfide S-(3-amino-3-carboxypropyl)transferase; O-succinyl-L-homoserine succinate-lyase (adding hydrogen sulfide) |
| R01777 | succinyl-CoA:L-homoserine O-succinyltransferase |
| R01851 | methanethiol:oxygen oxidoreductase |
| R01930 | trithionate thiosulfohydrolase |
| R01931 | thiosulfate:cyanide sulfurtranserase |
| R02021 | adenosine 3',5'-bisphosphate,sulfite:oxidized-thioredoxin oxidoreductase (3'-phosphoadenosine-5'-phosphosulfate -forming) |
| R02574 | S,S-dimethyl-beta-propiothetin dimethyl-sulfide-lyase |
| R03533 | donor:sulfur oxidoreductase |
| R05320 | taurine,2-oxoglutarate:O2 oxidoreductase (sulfite-forming) |
| R05717 | AMP,sulfite:glutathione-disulfide oxidoreductase (adenosine-5'-phosphosulfate-forming) |
| R07177 | thiosulfate:6-decylubiquinone oxidoreductase |
| R07210 | alkanesulfonate,reduced-FMN:oxygen oxidoreductase |
| R07365 | sulfur:oxygen oxidoreductase (hydrogen-sulfide- and sulfite-forming) |
| R08553 | AMP,sulfite:acceptor oxidoreductase (adenosine-5'-phosphosulfate-forming) |
| R08678 | S-sulfanylglutathione:oxygen oxidoreductase |
| R09498 | dimethylsulfone reductase |
| R09499 | hydrogen-sulfide:flavocytochrome c oxidoreductase |
| R09500 | dimethyl sulfide:ferricytochrome-c2 oxidoreductase |
| R09501 | dimethyl sulfide:menaquinone oxidoreductase |
| R09513 | methanesulfonate,NADH:oxygen oxidoreductase |
| R09786 | dimethyl sulfide,NADH:oxygen oxidoreductase |
| R09997 | methanethiol:coenzyme M methyltransferase |
| R10152 | sulfide:quinone oxidoreductase |
| R10206 | methanesulfonate,FMNH2:oxygen oxidoreductase |
| R10333 | S,S-dimethyl-beta-propiothetin:tetrahydrofolate S-methyltransferase |
| R10390 | H2:polysulfide oxidoreductase |
| R10419 | hydrogen sulfide:NAD+ oxidoreductase (CoA-dependent) |
| R10438 | hydrogen sulfide:NADP+ oxidoreductase (CoA-dependent) |
| R10820 | 3-(methylthio)propanoate:CoA ligase (AMP-forming) |
| R10936 | 3-(methylthio)acryloyl-CoA hydratase |
| R11130 | 3-(methylsulfanyl)propanoyl-CoA:acceptor 2-oxidoreductase |
| R11487 | sulfite:quinone oxidoreductase |
| R11546 | S-adenosyl-L-methionine:methanethiol S-methyltransferase |
| R11929 | sulfide:glutathione,quinone oxidoreductase |
| R12666 | S-adenosyl-L-methionine:(2R)-2-hydroxy-4-(methylsulfanyl)butanoate S-methyltransferase |
| R13227 | hydrogen sulfide:[DsrC protein]-dithiol oxidoreductase (trisulfide-forming) |
|
|---|
| Compound |
| C00053 | 3'-Phosphoadenylyl sulfate |
| C00054 | Adenosine 3',5'-bisphosphate |
| C01118 | O-Succinyl-L-homoserine |
| C01180 | 4-Methylthio-2-oxobutanoic acid |
| C03920 | 2-(Methylthio)ethanesulfonate |
| C04022 | S,S-Dimethyl-beta-propiothetin |
| C08276 | 3-(Methylthio)propanoate |
| C20870 | 3-(Methylthio)propanoyl-CoA |
| C20955 | 3-(Methylthio)acryloyl-CoA |
| C22315 | (2R)-2-Hydroxy-4-(methylsulfanyl)butanoate |
| C22316 | (2R)-4-(Dimethylsulfaniumyl)-2-hydroxybutanoate |
| C23001 | 3-Aminopropyl(dimethyl)sulfanium |
| C23002 | 3-Dimethylsulfoniopropionaldehyde |
|
|---|
| Reference |
|
|---|
| Authors |
Grein F, Ramos AR, Venceslau SS, Pereira IA |
|---|
| Title |
Unifying concepts in anaerobic respiration: Insights from dissimilatory sulfur metabolism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Fauque GD, Barton LL |
|---|
| Title |
Hemoproteins in dissimilatory sulfate- and sulfur-reducing prokaryotes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sakurai H, Ogawa T, Shiga M, Inoue K |
|---|
| Title |
Inorganic sulfur oxidizing system in green sulfur bacteria. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Falkenby LG, Szymanska M, Holkenbrink C, Habicht KS, Andersen JS, Miller M, Frigaard NU |
|---|
| Title |
Quantitative proteomics of Chlorobaculum tepidum: insights into the sulfur metabolism of a phototrophic green sulfur bacterium. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Gregersen LH, Bryant DA, Frigaard NU |
|---|
| Title |
Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Beller HR, Chain PS, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP. |
|---|
| Title |
The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pott AS, Dahl C |
|---|
| Title |
Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Frigaard NU, Dahl C |
|---|
| Title |
Sulfur metabolism in phototrophic sulfur bacteria. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00270 | Cysteine and methionine metabolism |
| rn00720 | Other carbon fixation pathways |
|
|---|
| KO pathway |
|
|---|