| Entry |
|
|---|
| Name |
Cysteine and methionine metabolism |
|---|
| Description |
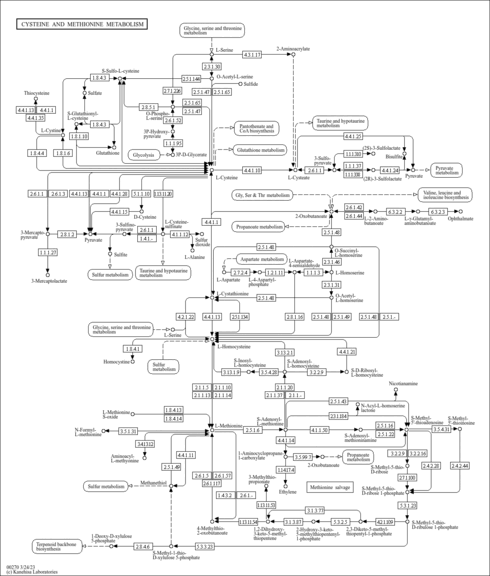
Cysteine and methionine are sulfur-containing amino acids. Cysteine is synthesized from serine through different pathways in different organism groups. In bacteria and plants, cysteine is converted from serine (via acetylserine) by transfer of hydrogen sulfide [MD: M00021]. In animals, methionine-derived homocysteine is used as sulfur source and its condensation product with serine (cystathionine) is converted to cysteine [MD: M00338]. Cysteine is metabolized to pyruvate in multiple routes. Methionine is an essential amino acid, which animals cannot synthesize. In bacteria and plants, methionine is synthesized from aspartate [MD: M00017]. S-Adenosylmethionine (SAM), synthesized from methionine and ATP, is a methyl group donor in many important transfer reactions including DNA methylation for regulation of gene expression. SAM may also be used to regenerate methionine in the methionine salvage pathway [MD: M00034].
|
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
| rn00270 | Cysteine and methionine metabolism |
|
|---|
| Module |
| M00017 | Methionine biosynthesis, aspartate => homoserine => methionine [PATH:rn00270] |
| M00338 | Cysteine biosynthesis, homocysteine + serine => cysteine [PATH:rn00270] |
| M00368 | Ethylene biosynthesis, methionine => ethylene [PATH:rn00270] |
| M00609 | Cysteine biosynthesis, methionine => cysteine [PATH:rn00270] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00075 | S-adenosyl-L-methionine:S-adenosyl-L-methionine 3-amino-3-carboxypropyltransferase |
| R00177 | ATP:L-methionine S-adenosyltransferase |
| R00178 | S-adenosyl-L-methionine carboxy-lyase [S-adenosyl 3-(methylsulfanyl)propylamine-forming] |
| R00179 | S-adenosyl-L-methionine methylthioadenosine-lyase (1-aminocyclopropane-1-carboxylate-forming) |
| R00192 | S-adenosyl-L-homocysteine hydrolase |
| R00193 | S-adenosyl-L-homocysteine aminohydrolase |
| R00194 | S-adenosyl-L-homocysteine homocysteinylribohydrolase |
| R00367 | S-adenosyl-L-methionine:glycine N-methyltransferase |
| R00480 | ATP:L-aspartate 4-phosphotransferase |
| R00586 | acetyl-CoA:L-serine O-acetyltransferase |
| R00648 | L-methionine:oxygen oxidoreductase (deaminating) |
| R00651 | O-acetyl-L-homoserine:methanethiol 3-amino-3-carboxypropyltransferase; O-acetyl-L-homoserine acetate-lyase (adding methanethiol) |
| R00653 | N-formyl-L-methionine amidohydrolase |
| R00654 | L-methionine methanethiol-lyase (deaminating;2-oxobutanoate-forming) |
| R00782 | L-cysteine hydrogen-sulfide-lyase (deaminating; pyruvate-forming) |
| R00863 | 3-sulfino-L-alanine 4-carboxy-lyase |
| R00892 | L-cysteine:NAD+ oxidoreductase; NADH:L-cystine oxidoreductase |
| R00893 | L-cysteine:oxygen oxidoreductase |
| R00895 | L-cysteine:2-oxoglutarate aminotransferase |
| R00896 | L-cysteine:2-oxoglutarate aminotransferase |
| R00897 | O3-acetyl-L-serine:hydrogen-sulfide 2-amino-2-carboxyethyltransferase; O3-acetyl-L-serine acetate-lyase (adding hydrogen sulfide) |
| R00900 | L-cysteine,glutathione:NADP+ oxidoreductase (disulfide-forming); NADPH:CoA-glutathione oxidoreductase |
| R00901 | L-cysteine hydrogen-sulfide-lyase (adding sulfite; L-cysteate-forming) |
| R00946 | 5-methyltetrahydrofolate:L-homocysteine S-methyltransferase |
| R00997 | 1-aminocyclopropane-1-carboxylate aminohydrolase (isomerizing); 1-aminocyclopropane-1-carboxylate endolyase (deaminating) |
| R00999 | O-succinyl-L-homoserine succinate-lyase (deaminating; 2-oxobutanoate-forming) |
| R01001 | L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming) |
| R01113 | glutathione:L-cystine oxidoreductase |
| R01286 | L-cystathionine L-homocysteine-lyase (deaminating; pyruvate-forming) |
| R01287 | O-acetyl-L-homoserine:hydrogen sulfide S-(3-amino-3-carboxypropyl)transferase; O-acetyl-L-homoserine acetate-lyase (adding hydrogen sulfide) |
| R01288 | O4-succinyl-L-homoserine:hydrogen sulfide S-(3-amino-3-carboxypropyl)transferase; O-succinyl-L-homoserine succinate-lyase (adding hydrogen sulfide) |
| R01290 | L-serine hydro-lyase (adding homocysteine; L-cystathionine-forming) |
| R01291 | S-(5-deoxy-D-ribos-5-yl)-L-homocysteine homocysteine-lyase [(4S)-4,5-dihydroxypentan-2,3-dione-forming] |
| R01292 | glutathione:homocystine oxidoreductase |
| R01401 | methylthioadenosine methylthioribohydrolase |
| R01402 | S-methyl-5'-thioadenosine:phosphate S-methyl-5-thio-alpha-D-ribosyl-transferase |
| R01513 | 3-phospho-D-glycerate:NAD+ 2-oxidoreductase |
| R01771 | ATP:L-homoserine O-phosphotransferase |
| R01773 | L-homoserine:NAD+ oxidoreductase |
| R01775 | L-homoserine:NADP+ oxidoreductase |
| R01776 | acetyl-CoA:L-homoserine O-acetyltransferase |
| R01777 | succinyl-CoA:L-homoserine O-succinyltransferase |
| R01874 | D-cysteine sulfide-lyase (deaminating; pyruvate-forming) |
| R01920 | S-adenosylmethioninamine:putrescine 3-aminopropyltransferase |
| R02025 | L-methionine:oxidized-thioredoxin S-oxidoreductase |
| R02026 | O-acetyl-L-homoserine acetate-lyase (L-homocysteine forming) |
| R02291 | L-aspartate-4-semialdehyde:NADP+ oxidoreductase (phosphorylating) |
| R02408 | L-cystine thiocysteine-lyase (deaminating; pyruvate-forming) |
| R02409 | coenzyme A:oxidized-glutathione oxidoreductase |
| R02433 | L-cysteate:2-oxoglutarate aminotransferase |
| R02619 | 3-sulfino-L-alanine:2-oxoglutarate aminotransferase |
| R02821 | trimethylaminoacetate:L-homosysteine S-methyltransferase |
| R02869 | S-adenosylmethioninamine:spermidine 3-aminopropyltransferase |
| R03104 | 3-mercaptolactate:NAD+ oxidoreductase |
| R03105 | 3-mercaptopyruvate:sulfide sulfurtransferase |
| R03132 | O-acetyl-L-serine:thiosulfate 2-amino-2-carboxyethyltransferase |
| R03217 | O-acetyl-L-homoserine succinate-lyase (adding cysteine) |
| R03260 | O-succinyl-L-homoserine succinate-lyase (adding cysteine) |
| R04143 | ATP:S5-methyl-5-thio-D-ribose 1-phosphotransferase |
| R04173 | 3-phosphoserine:2-oxoglutarate aminotransferase |
| R04405 | 5-methyltetrahydropteroyltri-L-glutamate:L-homocysteine S-methyltransferase |
| R04420 | 5-methylthio-5-deoxy-D-ribose-1-phosphate ketol-isomerase |
| R04858 | S-adenosyl-L-methionine:DNA (cytosine-5-)-methyltransferase |
| R05693 | (R)-2-hydroxyacid:NAD+ oxidoreductase |
| R07136 | (2R)-3-sulfolactate:NAD+ oxidoreductase |
| R07137 | (2R)-3-sulfolactate:NADP+ oxidoreductase |
| R07214 | 1-aminocyclopropane-1-carboxylate oxygenase (ethylene-forming) |
| R07274 | O-phospho-L-serine:hydrogen-sulfide 2-amino-2-carboxyethyltransferase |
| R07363 | 1,2-dihydroxy-5-(methylthio)pent-1-en-3-one:oxygen oxidoreductase (formate- and CO-forming) |
| R07364 | 1,2-dihydroxy-5-(methylthio)pent-1-en-3-one:oxygen oxidoreductase (formate-forming) |
| R07392 | S-methyl-5-thio-D-ribulose-1-phosphate hydro-lyase |
| R07393 | 2,3-diketo-5-methylthiopentyl-1-phosphate keto---enol-isomerase |
| R07394 | 2-hydroxy-5-(methylthio)-3-oxopent-1-enyl phosphate phosphohydrolase |
| R07395 | 5-(methylthio)-2,3-dioxopentyl-phosphate phosphohydrolase (isomerizing) |
| R07633 | 3-sulfolactate bisulfite-lyase |
| R07634 | L-cysteate bisulfite-lyase (deaminating) |
| R08699 | 3'-phosphoadenylyl sulfate:2-aminoacrylate C-sulfotransferase |
| R08940 | acyl-[acyl-carrier protein]:S-adenosyl-L-methionine acyltranserase (lactone-forming, methylthioadenosine-releasing) |
| R09660 | S-methyl-5'-thioadenosine amidohydrolase |
| R09668 | S-methyl-5'-thioinosine:phosphate S-methyl-5-thio-alpha-D-ribosyl-transferase |
| R10305 | O3-acetyl-L-serine:L-homocysteine S-(2-amino-2-carboxyethyl)transferase |
| R10991 | 2-aminobutanoate:2-oxoglutarate aminotransferase |
| R10992 | L-alanine:2-oxobutanoate aminotransferase |
| R10993 | L-glutamate:2-aminobutanoate gamma-ligase (ADP-forming) |
| R10994 | gamma-L-glutamyl-L-2-aminobutyrate:glycine ligase (ADP-forming) |
| R11063 | S-methyl-L-methionine:L-homocysteine S-methyltransferase |
| R12055 | L-methionine:indole-3-pyruvic acid aminotransferase |
| R12092 | thiosulfate:3-phospho-L-serine thiosulfotransferase |
| R12342 | ADP:L-serine 3-phosphotransferase |
| R12505 | hydrogen sulfide:L-aspartate-4-semialdehyde sulfurtransferase |
| R12507 | L-glutamine:4-(methylsulfanyl)-2-oxobutanoate aminotransferase |
| R12588 | S-inosyl-L-homocysteine hydrolase (inosine-forming) |
| R12718 | S-methyl-5-thio-D-ribulose 1-phosphate 1,3-isomerase |
| R12946 | S-methyl-1-thio-D-xylulose 5-phosphate:glutathione methylthiotransferase |
| R13363 | O-phospho-L-homoserine:L-cysteine S-(3-amino-3-carboxypropyl)transferase |
|
|---|
| Compound |
| C00019 | S-Adenosyl-L-methionine |
| C00021 | S-Adenosyl-L-homocysteine |
| C00441 | L-Aspartate 4-semialdehyde |
| C01118 | O-Succinyl-L-homoserine |
| C01137 | S-Adenosylmethioninamine |
| C01180 | 4-Methylthio-2-oxobutanoic acid |
| C01234 | 1-Aminocyclopropane-1-carboxylate |
| C03431 | S-Inosyl-L-homocysteine |
| C03539 | S-Ribosyl-L-homocysteine |
| C04188 | S-Methyl-5-thio-D-ribose 1-phosphate |
| C04582 | S-Methyl-5-thio-D-ribulose 1-phosphate |
| C05526 | S-Glutathionyl-L-cysteine |
| C08276 | 3-(Methylthio)propanoate |
| C11437 | 1-Deoxy-D-xylulose 5-phosphate |
| C15606 | 1,2-Dihydroxy-5-(methylthio)pent-1-en-3-one |
| C15650 | 2,3-Diketo-5-methylthiopentyl-1-phosphate |
| C15651 | 2-Hydroxy-3-keto-5-methylthiopentenyl-1-phosphate |
| C18049 | N-Acyl-L-homoserine lactone |
| C19787 | 5'-S-Methyl-5'-thioinosine |
| C21015 | gamma-L-Glutamyl-L-2-aminobutyrate |
| C22359 | S-Methyl-1-thio-D-xylulose 5-phosphate |
|
|---|
| Reference |
|
|---|
| Authors |
Sekowska A, Denervaud V, Ashida H, Michoud K, Haas D, Yokota A, Danchin A |
|---|
| Title |
Bacterial variations on the methionine salvage pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ashida H, Saito Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. |
|---|
| Title |
A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sekowska A, Danchin A. |
|---|
| Title |
The methionine salvage pathway in Bacillus subtilis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Berger BJ, English S, Chan G, Knodel MH. |
|---|
| Title |
Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Goyer A, Collakova E, Shachar-Hill Y, Hanson AD |
|---|
| Title |
Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rebeille F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S |
|---|
| Title |
Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pirkov I, Norbeck J, Gustafsson L, Albers E |
|---|
| Title |
A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ashida H, Saito Y, Kojima C, Yokota A |
|---|
| Title |
Enzymatic characterization of 5-methylthioribulose-1-phosphate dehydratase of the methionine salvage pathway in Bacillus subtilis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kitabatake M, So MW, Tumbula DL, Soll D |
|---|
| Title |
Cysteine biosynthesis pathway in the archaeon Methanosarcina barkeri encoded by acquired bacterial genes? |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mino K, Ishikawa K |
|---|
| Title |
Characterization of a novel thermostable O-acetylserine sulfhydrylase from Aeropyrum pernix K1. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Tanabe S |
|---|
| Title |
Development of assay methods for endogenous inorganic sulfur compounds and sulfurtransferases and evaluation of the physiological functions of bound sulfur. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL |
|---|
| Title |
Transition state analogs of 5'-methylthioadenosine nucleosidase disrupt quorum sensing. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00250 | Alanine, aspartate and glutamate metabolism |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00290 | Valine, leucine and isoleucine biosynthesis |
| rn00430 | Taurine and hypotaurine metabolism |
| rn00770 | Pantothenate and CoA biosynthesis |
| rn00900 | Terpenoid backbone biosynthesis |
|
|---|
| KO pathway |
|
|---|