| Entry |
|
|---|
| Name |
Biosynthesis of enediyne antibiotics |
|---|
| Description |
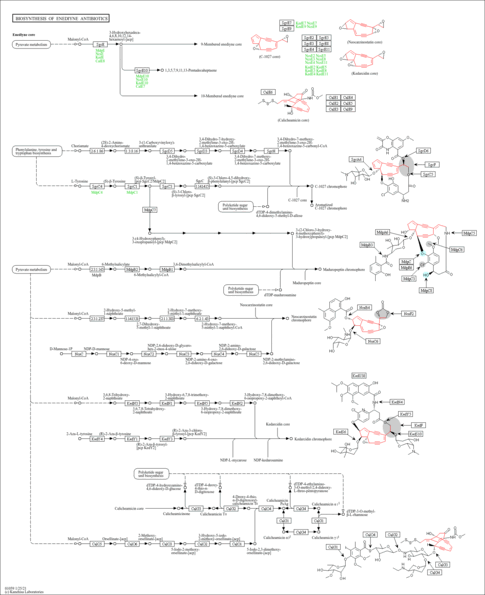
Enediyne natural products are potent antitumor antibiotics produced by a variety of Actinomycetes. Enediyne has a unique structure consisting of three building blocks: aromatic, sugar and enediyne core. The enediyne core contains two acetylenic groups conjugated to a double bond in a 9- or 10-membered ring, and it is synthesized by an iterative type I polyketide synthase and other tailoring proteins [MD: M00824 M00825]. The 10-membered enediyne such as calicheamicin also contains an allylic trisulfide group, which acts as a trigger for diradical formation. This diagram shows the biosynthesis of C-1027, maduropeptin, neocarzinostatin, kedarcidin and calicheamicin. Some aromatic moieties are synthesized via iterative type I polyketide synthases, and others are derived from chorismate and aromatic amino acids such as tyrosine and azatyrosine [MD: M00829 M00830 M00831 M00834 M00826 M00827 M00828 M00832]. Finally, the aromatic and sugar building blocks are attached to the enediyne core by acyltransferases, condensation enzymes and glucosyltransferases [MD: M00833].
|
|---|
| Class |
Metabolism; Metabolism of terpenoids and polyketides
|
|---|
| Pathway map |
| rn01059 | Biosynthesis of enediyne antibiotics |
|
|---|
| Module |
| M00824 | 9-membered enediyne core biosynthesis, malonyl-CoA => 3-hydroxyhexadeca-4,6,8,10,12,14-hexaenoyl-ACP => 9-membered enediyne core [PATH:rn01059] |
| M00825 | 10-membered enediyne core biosynthesis, malonyl-CoA => 3-hydroxyhexadeca-4,6,8,10,12,14-hexaenoyl-ACP => 10-membered enediyne core [PATH:rn01059] |
| M00826 | C-1027 benzoxazolinate moiety biosynthesis, chorismate => benzoxazolinyl-CoA [PATH:rn01059] |
| M00827 | C-1027 beta-amino acid moiety biosynthesis, tyrosine => 3-chloro-4,5-dihydroxy-beta-phenylalanyl-PCP [PATH:rn01059] |
| M00828 | Maduropeptin beta-hydroxy acid moiety biosynthesis, tyrosine => 3-(4-hydroxyphenyl)-3-oxopropanoyl-PCP [PATH:rn01059] |
| M00829 | 3,6-Dimethylsalicylyl-CoA biosynthesis, malonyl-CoA => 6-methylsalicylate => 3,6-dimethylsalicylyl-CoA [PATH:rn01059] |
| M00830 | Neocarzinostatin naphthoate moiety biosynthesis, malonyl-CoA => 2-hydroxy-5-methyl-1-naphthoate => 2-hydroxy-7-methoxy-5-methyl-1-naphthoyl-CoA [PATH:rn01059] |
| M00831 | Kedarcidin 2-hydroxynaphthoate moiety biosynthesis, malonyl-CoA => 3,6,8-trihydroxy-2-naphthoate => 3-hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoyl-CoA [PATH:rn01059] |
| M00832 | Kedarcidin 2-aza-3-chloro-beta-tyrosine moiety biosynthesis, azatyrosine => 2-aza-3-chloro-beta-tyrosyl-PCP [PATH:rn01059] |
| M00833 | Calicheamicin biosynthesis, calicheamicinone => calicheamicin [PATH:rn01059] |
| M00834 | Calicheamicin orsellinate moiety biosynthesis, malonyl-CoA => orsellinate-ACP => 5-iodo-2,3-dimethoxyorsellinate-ACP [PATH:rn01059] |
|
|---|
| Reaction |
| R07253 | acyl-CoA:malonyl-CoA C-acyltransferase (decarboxylating, oxoacyl-reducing, thioester-hydrolysing and cyclizing) |
| R08956 | (2S)-2-amino-4-deoxychorismate:L-glutamate aminotransferase |
| R09559 | (2S)-2-amino-4-deoxychorismate:FMN oxidoreductase |
| R10731 | (3S)-3-amino-3-(3-chloro-4-hydroxyphenyl)propanoyl-[peptidyl-carrier protein],FADH2:oxygen oxidoreductase (5-hydroxylating) |
| R10771 | 2-hydroxy-7-methoxy-5-methyl-1-naphthoate:CoA ligase |
| R10796 | 2-hydroxy-5-methyl-1-naphthoate,reduced ferredoxin:oxygen oxidoreductase (7-hydroxylating) |
| R10963 | S-adenosyl-L-methionine:2,7-dihydroxy-5-methyl-1-naphthoate 7-O-methyltransferase |
| R11125 | malonyl-CoA:acetyl-CoA malonyltransferase (2-hydroxy-5-methyl-1-naphthoic acid forming) |
| R11366 | L-tyrosine 2,3-aminomutase |
| R11367 | (S)-beta-tyrosine:[peptidyl-carrier protein] ligase (AMP-forming) |
| R11368 | (S)-beta-tyrosyl-[pcp]:FADH2 oxidoreductase |
| R11370 | 6-methylsalicylate:CoA ligase (AMP-forming) |
| R11371 | S-adenosyl-L-methionine:6-methylsalicylyl-CoA 3-C-methyltransferase |
| R11391 | 2-aza-L-tyrosine 2,3-aminomutase |
| R11392 | (R)-2-aza-beta-tyrosine:[peptidyl-carrier protein] ligase (AMP-forming) |
| R11393 | (R)-2-aza-beta-tyrosyl-[pcp]:FADH2 oxidoreductase |
| R11413 | S-adenosyl-L-methionine:orsellinate-[acp] O-methyltransferase |
| R11416 | S-adenosyl-L-methionine:3-hydroxy-5-iodo-2-methoxyorsellinate-[acp] O-methyltransferase |
| R11417 | dTDP-4-hydroxyamino-4,6-dideoxy-alpha-D-glucose:calicheamicinone 4-hydroxyamino-4,6-dideoxy-alpha-D-glucosyltransferase |
| R11418 | dTDP-4-deoxy-4-thio-alpha-D-digitoxose:calicheamicin-T0 4-deoxy-4-thio-alpha-D-digitoxosyltransferase |
| R11420 | dTDP-4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentopyranose:calicheamicin-PsAg 4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentosyltransferase |
| R11421 | dTDP-4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentopyranose:calicheamicin-alpha3(I) 4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentosyltransferase |
| R11422 | dTDP-3'-O-methyl-beta-L-rhamnose:calicheamicin-alpha1(I) 3'-O-methyl-beta-L-rhamnosyltransferase |
| R11423 | dTDP-3'-O-methyl-beta-L-rhamnose:calicheamicin-PsAg 3'-O-methyl-beta-L-rhamnosyltransferase |
|
|---|
| Compound |
| C11442 | Aromatized C-1027 chromophore |
| C11447 | dTDP-4-dimethylamino-4,6-dideoxy-5-C-methyl-D-allose |
| C11448 | 3,4-Dihydro-7-methoxy-2-methylene-3-oxo-2H-1,4-benzoxazine-5-carbonyl-CoA |
| C11449 | (S)-3-Chloro-4,5-dihydroxy-beta-phenylalanyl-[pcp] |
| C11468 | 3,4-Dihydro-7-methoxy-2-methylene-3-oxo-2H-1,4-benzoxazine-5-carboxylic acid |
| C11469 | Calicheamicin gamma(1)I |
| C12049 | Neocarzinostatin chromophore |
| C18054 | 2-Amino-2-deoxyisochorismate |
| C19686 | 3-(1-Carboxyvinyloxy)anthranilate |
| C20810 | (S)-3-Chloro-beta-tyrosyl-[pcp] |
| C20841 | 2-Hydroxy-7-methoxy-5-methyl-1-naphthoate |
| C20842 | 2-Hydroxy-7-methoxy-5-methyl-1-naphthoyl-CoA |
| C20856 | 2-Hydroxy-5-methyl-1-naphthoate |
| C20857 | 2,7-Dihydroxy-5-methyl-1-naphthoate |
| C21292 | Maduropeptin chromophore |
| C21306 | 3,4-Dihydro-2-methylene-3-oxo-2H-1,4-benzoxazine-5-carboxylate |
| C21307 | 3,4-Dihydro-7-hydroxy-2-methylene-3-oxo-2H-1,4-benzoxazine-5-carboxylate |
| C21311 | 3-(4-Hydroxyphenyl)-3-oxopropanoyl-[pcp] |
| C21313 | 3,6-Dimethylsalicylyl-CoA |
| C21315 | NDP-4-oxo-6-deoxy-D-mannose |
| C21316 | NDP-2,6-dideoxy-D-glycero-hex-2-enos-4-ulose |
| C21317 | NDP-2-amino-4-oxo-2,6-dideoxy-D-galactose |
| C21318 | NDP-2-amino-2,6-dideoxy-D-galactose |
| C21319 | NDP-2-methylamino-2,6-dideoxy-D-galactose |
| C21320 | 3,6,8-Trihydroxy-2-naphthoate |
| C21321 | 3,6,7,8-Tetrahydroxy-2-naphthoate |
| C21322 | 3-Hydroxy-6,7,8-trimethoxy-2-naphthoate |
| C21323 | 3-Hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoate |
| C21324 | 3-Hydroxy-7,8-dimethoxy-6-isopropoxy-2-naphthoyl-CoA |
| C21326 | (R)-2-Aza-beta-tyrosine |
| C21327 | (R)-2-Aza-beta-tyrosyl-[pcp] |
| C21328 | (R)-2-Aza-3-chloro-beta-tyrosyl-[pcp] |
| C21339 | 4-Deoxy-4-thio-alpha-D-digitoxosyl-calicheamicin T0 |
| C21341 | Calicheamicin alpha1(I) |
| C21342 | Calicheamicin alpha3(I) |
| C21344 | 2-Methoxyorsellinate-[acp] |
| C21345 | 5-Iodo-2-methoxyorsellinate-[acp] |
| C21346 | 3-Hydroxy-5-iodo-2-methoxyorsellinate-[acp] |
| C21347 | 5-Iodo-2,3-dimethoxyorsellinate-[acp] |
| C21348 | dTDP-4-hydroxyamino-4,6-dideoxy-alpha-D-glucose |
| C21349 | dTDP-4-deoxy-4-thio-alpha-D-digitoxose |
| C21350 | dTDP-3-O-methyl-beta-L-rhamnose |
| C21356 | dTDP-4-ethylamino-3-O-methyl-2,4-dideoxy-L-threo-pentopyranose |
| C21357 | 3-Hydroxyhexadeca-4,6,8,10,12,14-hexaenoyl-[acp] |
| C21358 | 1,3,5,7,9,11,13-Pentadecaheptaene |
| C21361 | 3-(2-Chloro-3-hydroxy-4-methoxyphenyl)-3-hydroxypropanoyl-[pcp] |
|
|---|
| Reference |
|
|---|
| Authors |
Liang ZX |
|---|
| Title |
Complexity and simplicity in the biosynthesis of enediyne natural products. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Shen B |
|---|
| Title |
Biosynthesis of enediyne antitumor antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Liu W, Christenson SD, Standage S, Shen B |
|---|
| Title |
Biosynthesis of the enediyne antitumor antibiotic C-1027. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS |
|---|
| Title |
The calicheamicin gene cluster and its iterative type I enediyne PKS. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B |
|---|
| Title |
Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B |
|---|
| Title |
The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lohman JR, Huang SX, Horsman GP, Dilfer PE, Huang T, Chen Y, Wendt-Pienkowski E, Shen B |
|---|
| Title |
Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Horsman GP, Van Lanen SG, Shen B |
|---|
| Title |
Iterative type I polyketide synthases for enediyne core biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sun H, Kong R, Zhu D, Lu M, Ji Q, Liew CW, Lescar J, Zhong G, Liang ZX |
|---|
| Title |
Products of the iterative polyketide synthases in 9- and 10-membered enediyne biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B |
|---|
| Title |
A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Belecki K, Crawford JM, Townsend CA |
|---|
| Title |
Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: implications for the biosynthesis of enediyne core structures. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kotaka M, Kong R, Qureshi I, Ho QS, Sun H, Liew CW, Goh LP, Cheung P, Mu Y, Lescar J, Liang ZX |
|---|
| Title |
Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Belecki K, Townsend CA |
|---|
| Title |
Biochemical determination of enzyme-bound metabolites: preferential accumulation of a programmed octaketide on the enediyne polyketide synthase CalE8. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Horsman GP, Chen Y, Thorson JS, Shen B |
|---|
| Title |
Polyketide synthase chemistry does not direct biosynthetic divergence between 9- and 10-membered enediynes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chen X, Ji R, Jiang X, Yang R, Liu F, Xin Y |
|---|
| Title |
Iterative type I polyketide synthases involved in enediyne natural product biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lin S, Horsman GP, Chen Y, Li W, Shen B |
|---|
| Title |
Characterization of the SgcF epoxide hydrolase supporting an (R)-vicinal diol intermediate for enediyne antitumor antibiotic C-1027 biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Horsman GP, Lechner A, Ohnishi Y, Moore BS, Shen B |
|---|
| Title |
Predictive model for epoxide hydrolase-generated stereochemistry in the biosynthesis of nine-membered enediyne antitumor antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lin S, Horsman GP, Shen B |
|---|
| Title |
Characterization of the epoxide hydrolase NcsF2 from the neocarzinostatin biosynthetic gene cluster. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Lin S, Shen B |
|---|
| Title |
Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Christenson SD, Wu W, Spies MA, Shen B, Toney MD |
|---|
| Title |
Kinetic analysis of the 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Lin S, Dorrestein PC, Kelleher NL, Shen B |
|---|
| Title |
Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Dorrestein PC, Christenson SD, Liu W, Ju J, Kelleher NL, Shen B |
|---|
| Title |
Biosynthesis of the beta-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring beta-amino acyl-S-carrier protein intermediates. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lin S, Van Lanen SG, Shen B |
|---|
| Title |
Characterization of the two-component, FAD-dependent monooxygenase SgcC that requires carrier protein-tethered substrates for the biosynthesis of the enediyne antitumor antibiotic C-1027. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Van Lanen SG, Lin S, Horsman GP, Shen B |
|---|
| Title |
Characterization of SgcE6, the flavin reductase component supporting FAD-dependent halogenation and hydroxylation in the biosynthesis of the enediyne antitumor antibiotic C-1027. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lohman JR, Shen B |
|---|
| Title |
4-methylideneimidazole-5-one-containing aminomutases in enediyne biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ling J, Horsman GP, Huang SX, Luo Y, Lin S, Shen B |
|---|
| Title |
Enediyne antitumor antibiotic maduropeptin biosynthesis featuring a C-methyltransferase that acts on a CoA-tethered aromatic substrate. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Huang SX, Lohman JR, Huang T, Shen B |
|---|
| Title |
A new member of the 4-methylideneimidazole-5-one-containing aminomutase family from the enediyne kedarcidin biosynthetic pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
McCoy JG, Johnson HD, Singh S, Bingman CA, Lei IK, Thorson JS, Phillips GN Jr |
|---|
| Title |
Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Singh S, Nandurkar NS, Thorson JS |
|---|
| Title |
Characterization of the calicheamicin orsellinate C2-O-methyltransferase CalO6. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chang A, Singh S, Bingman CA, Thorson JS, Phillips GN Jr |
|---|
| Title |
Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| rn00523 | Polyketide sugar unit biosynthesis |
|
|---|
| KO pathway |
|
|---|