| Entry |
|
|---|
| Name |
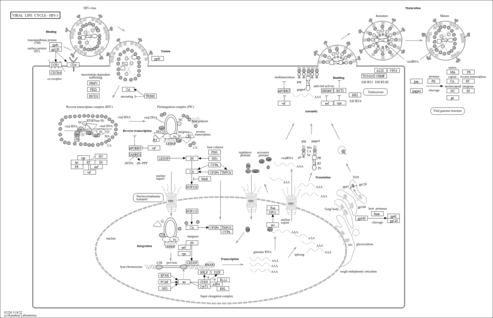
Viral life cycle - HIV-1 - Zea mays (maize) |
|---|
| Class |
Genetic Information Processing; Information processing in viruses
|
|---|
| Pathway map |
|
|---|
| Organism |
Zea mays (maize) [GN: zma]
|
|---|
| Gene |
|
|---|
| Reference |
|
|---|
| Authors |
Li G, De Clercq E |
|---|
| Title |
HIV Genome-Wide Protein Associations: a Review of 30 Years of Research. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Tagliamonte M, Tornesello ML, Buonaguro FM, Buonaguro L |
|---|
| Title |
Conformational HIV-1 envelope on particulate structures: a tool for chemokine coreceptor binding studies. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Novikova M, Zhang Y, Freed EO, Peng K |
|---|
| Title |
Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rawle DJ, Harrich D |
|---|
| Title |
Toward the "unravelling" of HIV: Host cell factors involved in HIV-1 core uncoating. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Le Sage V, Mouland AJ, Valiente-Echeverria F |
|---|
| Title |
Roles of HIV-1 capsid in viral replication and immune evasion. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Murphy RE, Saad JS |
|---|
| Title |
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
He N, Zhou Q |
|---|
| Title |
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pincetic A, Leis J |
|---|
| Title |
The Mechanism of Budding of Retroviruses From Cell Membranes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kleinpeter AB, Freed EO |
|---|
| Title |
HIV-1 Maturation: Lessons Learned from Inhibitors. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Young TM, Wang Q, Pe'ery T, Mathews MB |
|---|
| Title |
The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|