| Entry |
|
|---|
| Name |
|
|---|
| Formula |
C5H4N4O3
|
|---|
| Exact mass |
168.0283
|
|---|
| Mol weight |
168.11
|
|---|
| Structure |
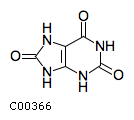
|
|---|
| Reaction |
|
|---|
| Pathway |
| map01120 | Microbial metabolism in diverse environments |
|
|---|
| Module |
| M00546 | Purine degradation, xanthine => urea |
| M00958 | Adenine ribonucleotide degradation, AMP => Urate |
| M00959 | Guanine ribonucleotide degradation, GMP => Urate |
|
|---|
| Network |
nt06025 Molybdenum cofactor biosynthesis nt06027 Purine salvage pathway |
|---|
| Enzyme |
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 12
1 C8y C 24.1500 -16.1000
2 C8y C 24.1500 -17.5000
3 C8y C 25.4100 -15.4000
4 N4x N 22.8900 -15.6800
5 N4x N 25.4100 -18.2000
6 N4x N 22.8900 -17.9200
7 N4x N 26.6000 -16.1000
8 O5x O 25.4100 -14.0000
9 C8y C 22.0500 -16.8000
10 C8y C 26.6000 -17.4300
11 O5x O 27.8600 -18.1300
12 O5x O 20.6500 -16.8000
BOND 13
1 1 2 2
2 1 3 1
3 1 4 1
4 2 5 1
5 2 6 1
6 3 7 1
7 3 8 2
8 4 9 1
9 5 10 1
10 10 11 2
11 6 9 1
12 7 10 1
13 9 12 2
|
|---|