| Entry |
|
|---|
| Name |
Oxidative phosphorylation - Arabidopsis thaliana (thale cress) |
|---|
| Class |
Metabolism; Energy metabolism
|
|---|
| Pathway map |
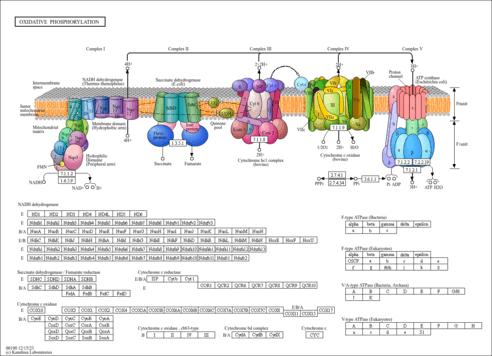
|
|---|
| Module |
| ath_M00143 | NADH dehydrogenase (ubiquinone) Fe-S protein/flavoprotein complex, mitochondria [PATH:ath00190] |
|
|---|
| Organism |
Arabidopsis thaliana (thale cress) [GN: ath]
|
|---|
| Gene |
| AT1G12840 | DET3; vacuolar ATP synthase subunit C (VATC) / V-ATPase C subunit / vacuolar proton pump C subunit (DET3) [KO:K02148] |
| AT1G75630 | AVA-P4; vacuolar H+-pumping ATPase 16 kDa proteolipid subunit 4 [KO:K02155] |
| AT4G32260 | PDE334; ATPase, F0 complex, subunit B/B', bacterial/chloroplast [KO:K02109] |
|
|---|
| Compound |
|
|---|
| Reference |
|
|---|
| Authors |
Sazanov LA, Hinchliffe P. |
|---|
| Title |
Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hinchliffe P, Carroll J, Sazanov LA. |
|---|
| Title |
Identification of a novel subunit of respiratory complex I from Thermus thermophilus. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|