| Entry |
|
|---|
| Name |
Propanoate metabolism - Sinorhizobium alkalisoli |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
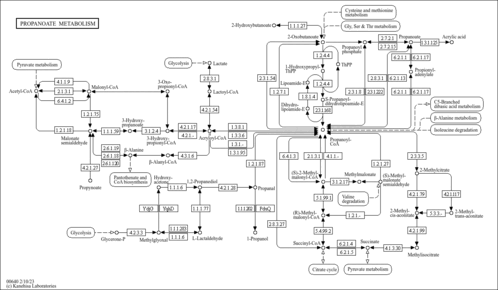
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Sinorhizobium alkalisoli [GN: eak]
|
|---|
| Gene |
|
|---|
| Compound |
| C04225 | (Z)-But-2-ene-1,2,3-tricarboxylate |
| C04593 | (2S,3R)-3-Hydroxybutane-1,2,3-tricarboxylate |
| C06002 | (S)-Methylmalonate semialdehyde |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C21017 | 2-(alpha-Hydroxypropyl)thiamine diphosphate |
| C21018 | Enzyme N6-(S-propyldihydrolipoyl)lysine |
| C21250 | 2-Methyl-trans-aconitate |
|
|---|
Related
pathway |
| eak00260 | Glycine, serine and threonine metabolism |
| eak00270 | Cysteine and methionine metabolism |
| eak00280 | Valine, leucine and isoleucine degradation |
| eak00660 | C5-Branched dibasic acid metabolism |
| eak00770 | Pantothenate and CoA biosynthesis |
|
|---|
| KO pathway |
|
|---|