| Entry |
|
|---|
| Name |
C5-Branched dibasic acid metabolism - Yersinia enterocolitica (type O:5) YE53/03 |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
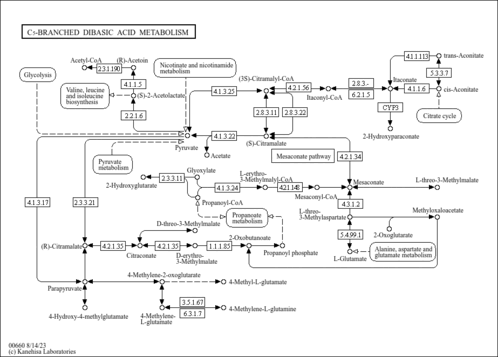
| yee00660 | C5-Branched dibasic acid metabolism |
|
|---|
| Organism |
Yersinia enterocolitica (type O:5) YE53/03 [GN: yee]
|
|---|
| Gene |
|
|---|
| Compound |
| C00651 | 4-Methylene-L-glutamate |
| C01109 | 4-Methylene-L-glutamine |
| C03618 | L-threo-3-Methylaspartate |
| C06027 | L-erythro-3-Methylmalyl-CoA |
| C06032 | D-erythro-3-Methylmalate |
| C06034 | 4-Hydroxy-4-methylglutamate |
| C06035 | 4-Methylene-2-oxoglutarate |
|
|---|
Related
pathway |
| yee00250 | Alanine, aspartate and glutamate metabolism |
| yee00290 | Valine, leucine and isoleucine biosynthesis |
| yee00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|