| Entry |
|
|---|
| Name |
IMP;
Inosinic acid;
Inosine monophosphate;
Inosine 5'-monophosphate;
Inosine 5'-phosphate;
5'-Inosinate;
5'-Inosinic acid;
5'-Inosine monophosphate;
5'-IMP
|
|---|
| Formula |
C10H13N4O8P
|
|---|
| Exact mass |
348.0471
|
|---|
| Mol weight |
348.21
|
|---|
| Structure |
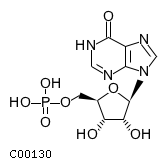
|
|---|
| Reaction |
|
|---|
| Pathway |
| map01060 | Biosynthesis of plant secondary metabolites |
| map01065 | Biosynthesis of alkaloids derived from histidine and purine |
| map01110 | Biosynthesis of secondary metabolites |
|
|---|
| Module |
| M00048 | De novo purine biosynthesis, PRPP + glutamine => IMP |
| M00049 | Adenine ribonucleotide biosynthesis, IMP => ADP,ATP |
| M00050 | Guanine ribonucleotide biosynthesis, IMP => GDP,GTP |
| M00958 | Adenine ribonucleotide degradation, AMP => Urate |
|
|---|
| Network |
|
|---|
| Enzyme |
|
|---|
| Brite |
Glycosides [BR:br08021]
N-glycosides
C00130 IMP
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 23
1 N4y N 25.9047 -15.8945
2 C8y C 24.5916 -15.4798
3 C1y C 24.8744 -17.9048
4 C8x C 26.7089 -14.7949
5 C8y C 24.5916 -14.0977
6 N5x N 23.3979 -16.1772
7 O2x O 23.7561 -17.0818
8 C1y C 24.4597 -19.1801
9 N5x N 25.9109 -13.6704
10 C8y C 23.3979 -13.4128
11 C8x C 22.2106 -15.4798
12 C1y C 22.6378 -17.8796
13 C1y C 23.0776 -19.1801
14 O1a O 25.2701 -20.2859
15 N4x N 22.2106 -14.0977
16 O5x O 23.3917 -12.0433
17 C1b C 21.3563 -17.4713
18 O1a O 22.2922 -20.2984
19 O2b O 20.3383 -18.0744
20 P1b P 18.9626 -18.0744
21 O1c O 17.5993 -18.0744
22 O1c O 18.9626 -16.7049
23 O1c O 18.9562 -19.4378
BOND 25
1 1 2 1
2 3 1 1 #Up
3 1 4 1
4 2 5 2
5 2 6 1
6 3 7 1
7 3 8 1
8 4 9 2
9 5 10 1
10 6 11 2
11 7 12 1
12 8 13 1
13 8 14 1 #Down
14 10 15 1
15 10 16 2
16 12 17 1 #Up
17 13 18 1 #Down
18 17 19 1
19 19 20 1
20 20 21 1
21 20 22 1
22 20 23 2
23 5 9 1
24 11 15 1
25 12 13 1
|
|---|