 | | PATHWAY: ecok00650 | |
| Entry |
|
|---|
| Name |
Butanoate metabolism - Escherichia coli K-12 MDS42 |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
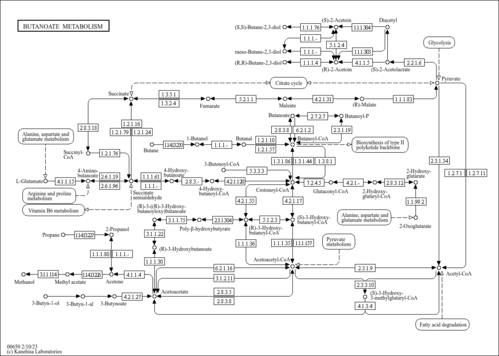
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Escherichia coli K-12 MDS42 [GN: ecok]
|
|---|
| Gene |
| ECMDS42_0571 | sdhC; succinate dehydrogenase, membrane subunit, binds cytochrome b556 [KO:K00241] |
| ECMDS42_0572 | sdhD; succinate dehydrogenase, membrane subunit, binds cytochrome b556 [KO:K00242] |
| ECMDS42_1367 | ydiF; fused predicted acetyl-CoA:acetoacetyl-CoA transferase: alpha subunit/beta subunit [KO:K19709] [EC:2.8.3.8] |
|
|---|
| Compound |
| C00356 | (S)-3-Hydroxy-3-methylglutaryl-CoA |
| C01144 | (S)-3-Hydroxybutanoyl-CoA |
| C03561 | (R)-3-Hydroxybutanoyl-CoA |
| C04546 | (R)-3-((R)-3-Hydroxybutanoyloxy)butanoate |
| C06143 | Poly-beta-hydroxybutyrate |
|
|---|
Related
pathway |
| ecok00250 | Alanine, aspartate and glutamate metabolism |
|
|---|
| KO pathway |
|
|---|
|
|