 | | PATHWAY: cti00650 | |
| Entry |
|
|---|
| Name |
Butanoate metabolism - Cupriavidus taiwanensis |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
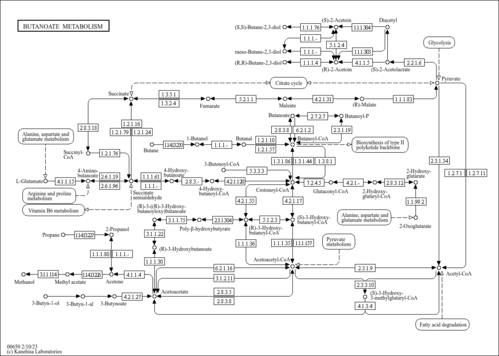
|
|---|
| Other DBs |
|
|---|
| Organism |
Cupriavidus taiwanensis [GN: cti]
|
|---|
| Gene |
| RALTA_B0510 | putative beta-ketoadipyl CoA thiolase with thiolase-like domain, phenylacetic acid degradation [KO:K00626] [EC:2.3.1.9] |
| RALTA_A1209 | putative nonspecific lipid-transfer protein, thiolase sterol carrier protein, ACETYL-COA C-ACETYLTRANSFERASE [KO:K00626] [EC:2.3.1.9] |
| RALTA_A1467 | fadB; multifunctional: 3-hydroxybutyryl-CoA epimerase, delta(3)-cis-delta(2)-trans-enoyl-CoA isomerase, enoyl-CoA hydratase (N-terminal); 3-hydroxyacyl-CoA dehydrogenase (C-terminal) [KO:K07516] [EC:1.1.1.35] |
| RALTA_A2131 | sdhC; succinate dehydrogenase, hydrophobic subunit, cytochrome b556 with SdhD [KO:K00241] |
| RALTA_A2130 | sdhD; succinate dehydrogenase, hydrophobic subunit, cytochrome b556 with SdhC [KO:K00242] |
| RALTA_A2344 | Acetoacetyl-coenzyme A synthetase (Acetoacetate--CoA ligase 1) (Acyl-activating enzyme 1) [KO:K01907] [EC:6.2.1.16] |
| RALTA_B0384 | putative Thiamine pyrophosphate-requiring enzyme; Benzoylformate decarboxylase, Acetolactate synthase isozyme II large subunit [KO:K01652] [EC:2.2.1.6] |
|
|---|
| Compound |
| C00356 | (S)-3-Hydroxy-3-methylglutaryl-CoA |
| C01144 | (S)-3-Hydroxybutanoyl-CoA |
| C03561 | (R)-3-Hydroxybutanoyl-CoA |
| C04546 | (R)-3-((R)-3-Hydroxybutanoyloxy)butanoate |
| C06143 | Poly-beta-hydroxybutyrate |
|
|---|
Related
pathway |
| cti00250 | Alanine, aspartate and glutamate metabolism |
| cti00330 | Arginine and proline metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|