| Entry |
|
|---|
| Name |
Alanine, aspartate and glutamate metabolism - Oryza sativa japonica (Japanese rice) (RAPDB) |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
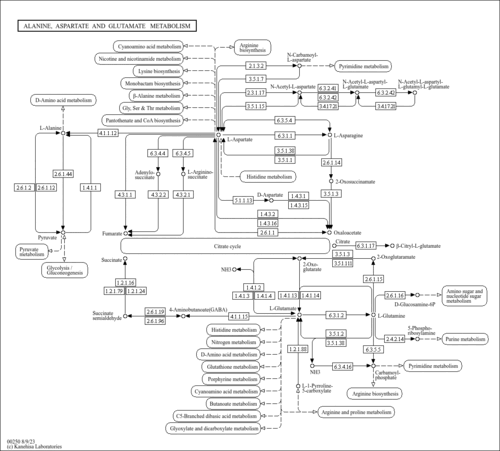
| dosa00250 | Alanine, aspartate and glutamate metabolism |
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Oryza sativa japonica (Japanese rice) (RAPDB) [GN: dosa]
|
|---|
| Gene |
| Os02t0134400-01 | (RAP-DB) Os02g0134400; Fumarate reductase/succinate dehydrogenase flavoprotein, C-terminal domain containing protein. [KO:K00278] [EC:1.4.3.16] |
| Os09t0433900-01 | (RAP-DB) Os09g0433900; Similar to Alanine aminotransferase 2 (EC 2.6.1.2) (GPT) (Glutamic--pyruvic transaminase 2) (Glutamic--alanine transaminase 2) (ALAAT-2). [KO:K00814] [EC:2.6.1.2] |
| Os10t0390600-01 | (RAP-DB) Os10g0390600; Similar to Alanine aminotransferase 2 (EC 2.6.1.2) (GPT) (Glutamic--pyruvic transaminase 2) (Glutamic--alanine transaminase 2) (ALAAT-2). [KO:K00814] [EC:2.6.1.2] |
| Os03t0699300-01 | (RAP-DB) Os03g0699300; Similar to Adenylosuccinate synthetase, chloroplast precursor (EC 6.3.4.4) (IMP-- aspartate ligase) (AdSS) (AMPSase). [KO:K01939] [EC:6.3.4.4] |
| Os03t0174500-01 | (RAP-DB) Os03g0174500; Similar to Adenylosuccinate synthetase, chloroplast precursor (EC 6.3.4.4) (IMP-- aspartate ligase) (AdSS) (AMPSase) (Fragment). [KO:K01939] [EC:6.3.4.4] |
| Os03t0313600-01 | (RAP-DB) Os03g0313600; Similar to Genes for GrpE, DnaK and DnaJ, complete and partial cds. (Fragment). [KO:K01756] [EC:4.3.2.2] |
| Os08t0248800-01 | (RAP-DB) Os08g0248800; Similar to Aspartate carbamoyltransferase 3, chloroplast precursor (EC 2.1.3.2) (Aspartate transcarbamylase 3) (ATCase 3). [KO:K00609] [EC:2.1.3.2] |
| Os04t0614500-01 | (RAP-DB) Os04g0614500; Pyridoxal phosphate-dependent transferase, major region, subdomain 1 domain containing protein. [KO:K16871] [EC:2.6.1.96] |
| Os01t0558200-01 | (RAP-DB) Os01g0558200; Glutamate/phenylalanine/leucine/valine dehydrogenase domain containing protein. [KO:K00262] [EC:1.4.1.4] |
| Os03t0223400-01 | (RAP-DB) Os03g0223400; Glutamine synthetase root isozyme (EC 6.3.1.2) (Glutamate--ammonia ligase) (Clone lambda-GS8). [KO:K01915] [EC:6.3.1.2] |
| Os02t0735200-01 | (RAP-DB) Os02g0735200; Glutamine synthetase shoot isozyme (EC 6.3.1.2) (Glutamate--ammonia ligase) (Clone lambda-GS28). [KO:K01915] [EC:6.3.1.2] |
| Os03t0712800-02 | (RAP-DB) Os03g0712800; Similar to Glutamine synthetase root isozyme 2 (EC 6.3.1.2) (Glutamate--ammonia ligase). [KO:K01915] [EC:6.3.1.2] |
| Os04t0659100-01 | (RAP-DB) Os04g0659100; Glutamine synthetase shoot isozyme, chloroplast precursor (EC 6.3.1.2) (Glutamate--ammonia ligase) (Clone lambda-GS31). [KO:K01915] [EC:6.3.1.2] |
| Os12t0131100-01 | (RAP-DB) Os12g0131100; Similar to Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] (EC 2.6.1.16) (Hexosephosphate aminotransferase) (D-fructose-6- phosphate amidotransferase) (GFAT). [KO:K00820] [EC:2.6.1.16] |
| Os05t0430800-01 | (RAP-DB) Os05g0430800; Similar to Amidophosphoribosyltransferase, chloroplast precursor (EC 2.4.2.14) (Glutamine phosphoribosylpyrophosphate amidotransferase) (ATASE) (GPAT). [KO:K00764] [EC:2.4.2.14] |
|
|---|
| Compound |
| C00352 | D-Glucosamine 6-phosphate |
| C00438 | N-Carbamoyl-L-aspartate |
| C03406 | N-(L-Arginino)succinate |
| C03794 | N6-(1,2-Dicarboxyethyl)-AMP |
| C03912 | (S)-1-Pyrroline-5-carboxylate |
| C12270 | N-Acetylaspartylglutamate |
| C20775 | beta-Citryl-L-glutamate |
| C20776 | N-Acetylaspartylglutamylglutamate |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Wu G |
|---|
| Title |
Intestinal mucosal amino acid catabolism. |
|---|
| Journal |
|
|---|
Related
pathway |
| dosa00260 | Glycine, serine and threonine metabolism |
| dosa00520 | Amino sugar and nucleotide sugar metabolism |
| dosa00630 | Glyoxylate and dicarboxylate metabolism |
| dosa00660 | C5-Branched dibasic acid metabolism |
| dosa00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|