| Entry |
|
|---|
| Name |
Propanoate metabolism - Mycobacterium tuberculosis variant bovis AF2122/97 |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
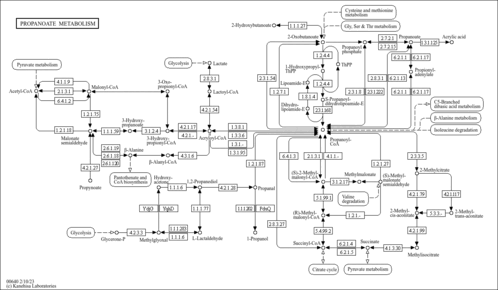
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Mycobacterium tuberculosis variant bovis AF2122/97 [GN: mbo]
|
|---|
| Gene |
| BQ2027_MB3691 | acs; ACETYL-COENZYME A SYNTHETASE ACS (ACETATE--CoA LIGASE) (ACETYL-CoA SYNTHETASE) (ACETYL-CoA SYNTHASE) (ACYL-ACTIVATING ENZYME) (ACETATE THIOKINASE) (ACETYL-ACTIVATING ENZYME) (ACETATE--COENZYME A LIGASE) (ACETYL-COENZYME A SYNTHASE) [KO:K01895] [EC:6.2.1.1] |
| BQ2027_MB0471 | lpd; dihydrolipoamide dehydrogenase lpdc (lipoamide reductase (nadh)) (lipoyl dehydrogenase) (dihydrolipoyl dehydrogenase) (diaphorase) [KO:K00382] [EC:1.8.1.4] |
| BQ2027_MB1099C | echA8; PROBABLE ENOYL-CoA HYDRATASE ECHA8 (ENOYL HYDRASE) (UNSATURATED ACYL-CoA HYDRATASE) (CROTONASE) [KO:K01692] [EC:4.2.1.17] |
| BQ2027_MB2511 | echA14; PROBABLE ENOYL-COA HYDRATASE ECHA14 (ENOYL HYDRASE) (UNSATURATED ACYL-COA HYDRATASE) (CROTONASE) [KO:K01692] [EC:4.2.1.17] |
| BQ2027_MB1507 | echA12; POSSIBLE ENOYL-CoA HYDRATASE ECHA12 (ENOYL HYDRASE) (UNSATURATED ACYL-CoA HYDRATASE) (CROTONASE) [KO:K01692] [EC:4.2.1.17] |
| BQ2027_MB3065C | echA17; PROBABLE ENOYL-COA HYDRATASE ECHA17 (CROTONASE) (UNSATURED ACYL-CoA HYDRATASE) (ENOYL HYDRASE) [KO:K01692] [EC:4.2.1.17] |
| BQ2027_MB3313 | accA3; PROBABLE BIFUNCTIONAL PROTEIN ACETYL-/PROPIONYL-COENZYME A CARBOXYLASE (ALPHA CHAIN) ACCA3: BIOTIN CARBOXYLASE + BIOTIN CARBOXYL CARRIER PROTEIN (BCCP) [KO:K11263] [EC:6.4.1.2 6.4.1.3 6.3.4.14] |
| BQ2027_MB2620 | gabT; 4-AMINOBUTYRATE AMINOTRANSFERASE GABT (GAMMA-AMINO-N-BUTYRATE TRANSAMINASE) (GABA TRANSAMINASE) (GLUTAMATE:SUCCINIC SEMIALDEHYDE TRANSAMINASE) (GABA AMINOTRANSFERASE) (GABA-AT) [KO:K07250] [EC:2.6.1.19 2.6.1.22 2.6.1.48] |
| BQ2027_MB3309 | acce5; probable bifunctional protein acetyl-/propionyl-coenzyme a carboxylase (epsilon chain) acce5 [KO:K27095] |
|
|---|
| Compound |
| C04225 | (Z)-But-2-ene-1,2,3-tricarboxylate |
| C04593 | (2S,3R)-3-Hydroxybutane-1,2,3-tricarboxylate |
| C06002 | (S)-Methylmalonate semialdehyde |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C21017 | 2-(alpha-Hydroxypropyl)thiamine diphosphate |
| C21018 | Enzyme N6-(S-propyldihydrolipoyl)lysine |
| C21250 | 2-Methyl-trans-aconitate |
|
|---|
Related
pathway |
| mbo00260 | Glycine, serine and threonine metabolism |
| mbo00270 | Cysteine and methionine metabolism |
| mbo00280 | Valine, leucine and isoleucine degradation |
| mbo00660 | C5-Branched dibasic acid metabolism |
| mbo00770 | Pantothenate and CoA biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|